A complex interplay of genetic introgression and local adaptation during the evolutionary history of three closely related spruce species
- PMID: 40734838
- PMCID: PMC12302640
- DOI: 10.1016/j.pld.2025.04.007
A complex interplay of genetic introgression and local adaptation during the evolutionary history of three closely related spruce species
Abstract
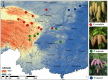
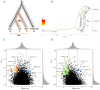
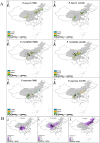
As climate change triggers unprecedented ecological shifts, it becomes imperative to understand the genetic underpinnings of species' adaptability. Adaptive introgression significantly contributes to organismal adaptation to new environments by introducing genetic variation across species boundaries. However, despite growing recognition of its importance, the extent to which adaptive introgression has shaped the evolutionary history of closely related species remains poorly understood. Here we employed population genetic analyses of high-throughput sequencing data to investigate the interplay between genetic introgression and local adaptation in three species of spruce trees in the genus Picea (P. asperata, P. crassifolia, and P. meyeri). We find distinct genetic differentiation among these species, despite a substantial gene flow. Crucially, we find bidirectional adaptive introgression between allopatrically distributed species pairs and unearthed dozens of genes linked to stress resilience and flowering time. These candidate genes most likely have promoted adaptability of these spruces to historical environmental changes and may enhance their survival and resilience to future climate changes. Our findings highlight that adaptive introgression could be prevalent and bidirectional in a topographically complex area, and this could have contributed to rich genetic variation and diverse habitat usage by tree species.
Keywords: Adaptation; Introgression; Picea; Population transcriptome.
© 2025 Kunming Institute of Botany, Chinese Academy of Sciences. Publishing services by Elsevier B.V. on behalf of KeAi Communications Co., Ltdé.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

Similar articles
-
Adaptive Introgression as an Evolutionary Force: A Meta-Analysis of Knowledge Trends.Evol Appl. 2025 Jun 20;18(6):e70103. doi: 10.1111/eva.70103. eCollection 2025 Jun. Evol Appl. 2025. PMID: 40548230 Free PMC article.
-
"In a State of Flow": A Qualitative Examination of Autistic Adults' Phenomenological Experiences of Task Immersion.Autism Adulthood. 2024 Sep 16;6(3):362-373. doi: 10.1089/aut.2023.0032. eCollection 2024 Sep. Autism Adulthood. 2024. PMID: 39371355
-
Genomic investigations of successful invasions: the picture emerging from recent studies.Biol Rev Camb Philos Soc. 2025 Jun;100(3):1396-1418. doi: 10.1111/brv.70005. Epub 2025 Feb 16. Biol Rev Camb Philos Soc. 2025. PMID: 39956989 Free PMC article.
-
Evolutionary Genomics Unravels the Responses and Adaptation to Climate Change in a Key Alpine Forest Tree Species.Mol Biol Evol. 2025 Jul 1;42(7):msaf116. doi: 10.1093/molbev/msaf116. Mol Biol Evol. 2025. PMID: 40415256 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
References
-
- Abdirad S., Wu Y.Q., Ghorbanzadeh Z., et al. Proteomic analysis of the meristematic root zone in contrasting genotypes reveals new insights in drought tolerance in rice. Proteomics. 2022;22 - PubMed
-
- Amraee L., Rahmani F. Modified CTAB protocol for RNA extraction from Lemon balm (Melissa officinalis L.) Acta Agric. Slov. 2020;115:53–57.
-
- Barton N.H. Speciation. Trends Ecol. Evol. 2001;16:325. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

