Troponin I - a comprehensive review of its function, structure, evolution, and role in muscle diseases
- PMID: 40735528
- PMCID: PMC12305882
- DOI: 10.1080/19768354.2025.2533821
Troponin I - a comprehensive review of its function, structure, evolution, and role in muscle diseases
Abstract
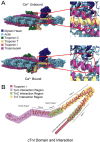
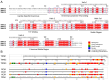
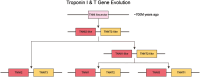
The troponin complex is a critical component of thin filaments and plays an essential role in the calcium-mediated regulation of contraction and relaxation in striated muscles, including both cardiac and skeletal muscle. Troponin I, a subunit of this complex, inhibits actomyosin interactions during muscle relaxation. Its function is finely tuned by posttranslational modifications, particularly phosphorylation, which influence calcium sensitivity and actin affinity, thus impacting muscle contraction. Mutations in troponin I are closely associated with various human diseases. Specifically, several mutations in cardiac troponin I have been linked to cardiomyopathies, such as hypertrophic, dilated, and restrictive cardiomyopathies, which affect heart contractility and calcium handling. In this review, we explore the multifaceted aspects of troponin I, including its structure, functional role in muscle contraction, evolution, and the complex interactions between posttranslational modifications and genetic mutations that alter its function and contribute to disease progression.
Keywords: Troponin I; cardiomyopathy; evolution of troponin I; genetic mutations in troponin I; post-transcriptional modification of troponin I.
© 2025 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
Similar articles
-
Velcro-binding by cardiac troponin-I traps tropomyosin on actin in a low-energy relaxed state.Biochem Biophys Res Commun. 2025 Apr 9;757:151595. doi: 10.1016/j.bbrc.2025.151595. Epub 2025 Mar 8. Biochem Biophys Res Commun. 2025. PMID: 40088678
-
Dual role of Tropomyosin-R160 in thin filament regulation: Insights into phosphorylation-dependent cardiac relaxation and cardiomyopathy mechanisms.Arch Biochem Biophys. 2025 Jun;768:110380. doi: 10.1016/j.abb.2025.110380. Epub 2025 Mar 6. Arch Biochem Biophys. 2025. PMID: 40057222
-
The highly conserved C-terminal end segment of troponin T binds tropomyosin and actin to function in modulating contractile kinetics.Proc Natl Acad Sci U S A. 2025 Jul 8;122(27):e2507107122. doi: 10.1073/pnas.2507107122. Epub 2025 Jul 1. Proc Natl Acad Sci U S A. 2025. PMID: 40591592
-
Management of urinary stones by experts in stone disease (ESD 2025).Arch Ital Urol Androl. 2025 Jun 30;97(2):14085. doi: 10.4081/aiua.2025.14085. Epub 2025 Jun 30. Arch Ital Urol Androl. 2025. PMID: 40583613 Review.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
References
-
- Alejandra Restrepo-Cordoba M, Campuzano O, Ripoll-Vera T, Cobo-Marcos M, Mademont-Soler I, Gamez JM, Dominguez F, Gonzalez-Lopez E, Padron-Barthe L, Lara-Pezzi E, et al. 2017. Usefulness of genetic testing in hypertrophic cardiomyopathy: an analysis using real-world data. J Cardiovasc Transl Res. 10:35–46. doi: 10.1007/s12265-017-9730-8. - DOI - PubMed
-
- Alfares AA, Kelly MA, McDermott G, Funke BH, Lebo MS, Baxter SB, Shen J, McLaughlin HM, Clark EH, Babb LJ, et al. 2015. Results of clinical genetic testing of 2,912 probands with hypertrophic cardiomyopathy: expanded panels offer limited additional sensitivity. Genet Med. 17:880–888. doi: 10.1038/gim.2014.205. - DOI - PubMed
-
- Alimohamed MZ, Johansson LF, Posafalvi A, Boven LG, van Dijk KK, Walters L, Vos YJ, Westers H, Hoedemaekers YM, Sinke RJ, et al. 2021. Diagnostic yield of targeted next generation sequencing in 2002 Dutch cardiomyopathy patients. Int J Cardiol. 332:99–104. doi: 10.1016/j.ijcard.2021.02.069. - DOI - PubMed
-
- Al-Shafai KN, Al-Hashemi M, Manickam C, Musa R, Selvaraj S, Syed N, Vempalli F, Ali M, Yacoub M, Estivill X.. 2021. Genetic evaluation of cardiomyopathies in Qatar identifies enrichment of pathogenic sarcomere gene variants and possible founder disease mutations in the Arabs. Mol Genet Genomic Med. 9:e1709. doi: 10.1002/mgg3.1709. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
