The nrfA-type microbial communities are widespread in hot springs of the Tibet-Yunnan geothermal zone
- PMID: 40735617
- PMCID: PMC12303954
- DOI: 10.3389/fmicb.2025.1540611
The nrfA-type microbial communities are widespread in hot springs of the Tibet-Yunnan geothermal zone
Abstract
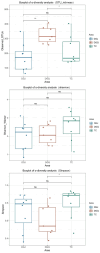
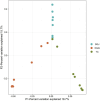
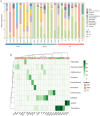
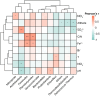
The microorganisms are main drivers of biogeochemical processes in geothermal ecosystems. The dissimilatory nitrate-to-ammonium reduction pathway (DNRA) could act as an alternative source of ammonium and provide an important nitrogen supply for the maintenance of geothermal ecosystems. Investigating the distribution of DNRA-functional bacteria is of great significance to understanding the source of biological nitrogen production in geothermal environments. In this study, we characterized the community distribution of microorganisms harboring nrfA genes in the sediments of hot springs from the Tibet-Yunnan geothermal zone, with the use of Illumina MiSeq high-throughput sequencing of nrfA genes and R language software for statistical analysis. In the present study, the nrfA genes were successfully amplified from the hot springs with a temperature of 38°C-80°C. The nrfA-based phylogenetic analysis showed that the DNRA pathway is widespread within the geothermal ecosystems, with microorganisms harboring nrfA genes predominantly belonging to phyla Chloroflexi, Proteobacteria, Deinococcus-Thermus (top 10), etc. Genus-level analysis revealed Thermoflexus (Chloroflexi) as the dominant taxon in the DGQ, while Geothrix (Acidobacteria) showed peak abundance in weakly acidic sites. The DNRA-functional community structure and nrfA gene abundance also showed a sample variability, even among samples from the same region, there were differences in dominant populations and overall nrfA gene abundance between them. Statistical analysis results indicate that the distribution of nrfA type microorganisms was mainly influenced by physicochemical factors, including pH, SO4 2-, and NO2 - concentrations. These findings deepen our understanding of the nitrogen cycle in extreme environments and provide valuable perspectives on the role of nitrogen metabolism in both contemporary and ancient geothermal systems.
Keywords: DNRA; Tibetan-Yunnan geothermal zone; hot springs; microbial community; nrfA gene.
Copyright © 2025 Chen, Wu, Yu, Li and Jiang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







Similar articles
-
Impact of seasonal change on dissimilatory nitrate reduction to ammonium (DNRA) triggering the retention of nitrogen in lake.J Environ Manage. 2023 Sep 1;341:118050. doi: 10.1016/j.jenvman.2023.118050. Epub 2023 May 2. J Environ Manage. 2023. PMID: 37141713
-
Comparing microbial populations from diverse hydrothermal features in Yellowstone National Park: hot springs and mud volcanoes.Front Microbiol. 2024 Jun 27;15:1409664. doi: 10.3389/fmicb.2024.1409664. eCollection 2024. Front Microbiol. 2024. PMID: 38993494 Free PMC article.
-
Hydrogen-dependent dissimilatory nitrate reduction to ammonium enables growth of Campylobacterota isolates.ISME J. 2025 Jan 2;19(1):wraf092. doi: 10.1093/ismejo/wraf092. ISME J. 2025. PMID: 40367351 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2020 Jan 9;1(1):CD011535. doi: 10.1002/14651858.CD011535.pub3. Cochrane Database Syst Rev. 2020. Update in: Cochrane Database Syst Rev. 2021 Apr 19;4:CD011535. doi: 10.1002/14651858.CD011535.pub4. PMID: 31917873 Free PMC article. Updated.
References
-
- Bennett A. C., Murugapiran S. K., Hamilton T. L. (2020). Temperature impacts community structure and function of phototrophic Chloroflexi and Cyanobacteria in two alkaline hot springs in Yellowstone National Park. Environ. Microbiol. Rep. 12, 503–513. doi: 10.1111/1758-2229.12863, PMID: - DOI - PMC - PubMed
-
- Bernard R. J., Mortazavi B., Kleinhuizen A. A. (2015). Dissimilatory nitrate reduction to ammonium (DNRA) seasonally dominates NO3− reduction pathways in an anthropogenically impacted sub-tropical coastal lagoon. Biogeochemistry 125, 47–64. doi: 10.1007/s10533-015-0111-6 - DOI
LinkOut - more resources
Full Text Sources

