Cytomegalovirus latency-the sum of subtleties
- PMID: 40736257
- PMCID: PMC12363206
- DOI: 10.1128/jvi.00664-25
Cytomegalovirus latency-the sum of subtleties
Abstract
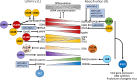
Human cytomegalovirus (HCMV) is a betaherpesvirus, which, like all herpesviruses, establishes a life-long latent infection while retaining the ability to reactivate its replicative program. While HCMV likely reactivates frequently and sporadically in healthy individuals and typically without disease, reactivation poses a serious disease threat in the immunocompromised. The latent program of HCMV is complex and has been challenging to define due to limitations in appropriate experimental model systems related to virus-host species specificity, limited identification of in vivo latent reservoirs, and the dynamic cellular differentiation of the hematopoietic latency reservoir that is directly linked to latency maintenance and reactivation phenotypes. Here, we review the current understanding of HCMV latency, with a focus on cross-cutting principles derived collectively from in vitro experimental culture models and in vivo animal models using the corresponding orthologs (CMVs) to HCMV.
Keywords: cytomegalovirus; herpesvirus; latency; virus-host interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Flint SJ, Racaniello VR, Rall GF, Skalka AM, Enquist LW. 2015. Principles of virology. 4th edition. ed. ASM Press, Washington, DC.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

