The P132H mutation of SARS-CoV-2 NSP5 relieves its inhibition on interferon-β activation via blocking MAVS degradation
- PMID: 40736574
- PMCID: PMC12311081
- DOI: 10.1007/s00018-025-05822-6
The P132H mutation of SARS-CoV-2 NSP5 relieves its inhibition on interferon-β activation via blocking MAVS degradation
Abstract
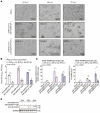
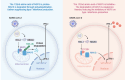
The prevalence of the Omicron variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is an important transition in the epidemic of coronavirus disease 2019 (COVID-19). Compared with other SARS-CoV-2 variants, Omicron and its subvariants exhibit decreased pathogenicity, thus contributing to the moderation of the epidemic. However, the mechanism underlying such changes is not fully understood. NSP5 is a SARS-CoV-2-encoded protease that counteracts antiviral immunity, and the P132H mutation of NSP5 is present exclusively in Omicron and its subvariants. In this study, we found that this mutation solely relieved cytopathogenicity and reduced the viral replication during SARS-CoV-2 infection. Further studies suggested that P132H blocked the NSP5-mediated degradation of MAVS by impairing the K136-linked ubiquitination of MAVS, thus restoring the IFN-β activation inhibited by NSP5. Structural analysis in silico suggested that P132H disrupted multiple hydrogen bonds between NSP5 and UbcH5b, an E2 ubiquitin-conjugating enzyme required for K136 ubiquitination. In summary, our results provide a potential mechanism explaining the decreased pathogenicity of the Omicron variant of SARS-CoV-2.
Keywords: IFN-β; MAVS; NSP5; P132H; SARS-CoV-2 variants.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors have no relevant financial or non-financial interests to disclose.
Figures


References
MeSH terms
Substances
Supplementary concepts
Grants and funding
- 32270170,81902070/National Natural Science Foundation of China
- 32322087,32300134/National Natural Science Foundation of China
- 2021YFC0866100, 2023YFC3041600/National Key Research and Development Program of China
- S2024JJMSXM2098/Provincial Natural Science Foundation of Hunan Province
- 2024RC1028/Science and Technology Innovation Program of Hunan Province
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

