Reconciling multiple connectivity-based systems biology methods for drug repurposing
- PMID: 40736744
- PMCID: PMC12309248
- DOI: 10.1093/bib/bbaf387
Reconciling multiple connectivity-based systems biology methods for drug repurposing
Abstract
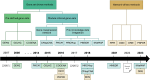
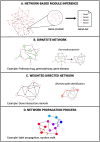
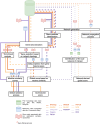
In the last two decades, numerous in silico methods have been developed for drug repurposing, to accelerate and reduce the risks about early drug development. Particularly, following Connectivity Map, dozens of distinct data-driven methods have been implemented to find candidates from the comparison of differential transcriptomic signatures. Interestingly, there have been multiple proposals to integrate available knowledge using systems biology databases and adapted algorithms from the network biology research field. Despite their similarities, these methods have been formulated inconsistently over the years, even if some of them are fundamentally similar. The aim of this review is to reconcile these integrative methods, focusing on elucidating their common structures while underlining the specificities of their strategies. To achieve this, we classified those methods into two main categories, provided schematic workflow representations, and presented a homogenized formulation for each.
Keywords: connectivity score; data integration; differential expression signature; drug repurposing; system biology.
© The Author(s) 2025. Published by Oxford University Press.
Figures
References
-
- Austin D, Hayford T, Kile J. et al. Research and Development in the Pharmaceutical Industry. Washington, DC: Congressional Budget Office; 2021 April. https://www.cbo.gov/publication/57025
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

