A comparative review of short genetic variant databases across humans and animal species
- PMID: 40736747
- PMCID: PMC12309242
- DOI: 10.1093/bib/bbaf356
A comparative review of short genetic variant databases across humans and animal species
Abstract
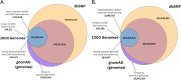
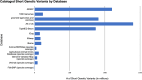
Understanding genetic variation is necessary to unravel the complexities of evolution and diverse traits across species. Short genetic variants (<50 bp in length) represent key genomic variations that play a crucial role in shaping the genetic landscape of populations. Human short genetic variant databases are often considered the gold standard for variant repositories, and this review compares them with variant databases created for various animal species. This review examines the methodologies, data integration, and various applications that differ or align between human and animal databases. The goal is to identify challenges in leveraging genetic variation across species, recommend strategies for overcoming these challenges, and suggest future directions for research and database development.
Keywords: allele frequencies; genetics; genomics; whole-genome sequencing.
© The Author(s) 2025. Published by Oxford University Press.
Figures
References
-
- Richards S, Aziz N, Bale S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–24. 10.1038/gim.2015.30 - DOI - PMC - PubMed
-
- Phasing out Support for Non-human Genome Organism Data in dbSNP and dbVar - NCBI Insights . https://ncbiinsights.ncbi.nlm.nih.gov/2017/05/09/phasing-out-support-for... (accessed Jul. 02, 2024).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

