Biomimetic targeted self-adaptive nanodrug for inflammation optimization and AT2 cell modulation in precise ARDS therapy
- PMID: 40737399
- PMCID: PMC12309668
- DOI: 10.1126/sciadv.adw5133
Biomimetic targeted self-adaptive nanodrug for inflammation optimization and AT2 cell modulation in precise ARDS therapy
Abstract
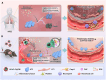
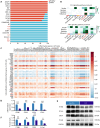
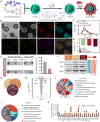
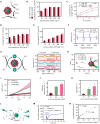
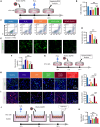
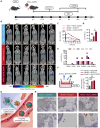
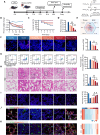
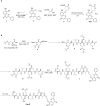
Acute respiratory distress syndrome (ARDS) is a lethal respiratory condition, while effective pharmacological treatments remain elusive. We identified the decreased mechanical capacity and impaired proliferation of alveolar type 2 (AT2) epithelial cells in the inflammatory environment as the primary contributors to respiratory failure of ARDS. A biomimetic, self-adaptive, 7,8-dihydroxyflavone-loaded hollow mesoporous cerium oxide coated with a platelet membrane (HCeOx-D@PM) was developed for precise ARDS therapy. HCeOx-D@PM comprises a platelet membrane (PM) shell for targeted delivery to injured lungs and an HCeOx core, which enables high drug loading, efficient reactive oxygen species (ROS) scavenging, and penetration of the alveolar-capillary barrier. Initially, HCeOx-D@PM suppresses the inflammation and mitigates the adverse effects of lesions on AT2 cell by scavenging accumulated ROS. It then adaptively releases 7,8-dihydroxyflavone in response to cysteine-aspartic acid protease 3 activation, facilitating AT2 cell proliferation and notably improving survival rates in vivo, offering a promising advancement in the precise treatment of respiratory diseases.
Figures

References
-
- Duggal A., Camporota L., Sequencing interventions in ARDS: The critical role of timing and order in standardized management. Intensive Care Med. 50, 1133–1136 (2024). - PubMed
-
- Bobba C. M., Fei Q., Shukla V., Lee H., Patel P., Putman R. K., Spitzer C., Tsai M., Wewers M. D., Lee R. J., Christman J. W., Ballinger M. N., Ghadiali S. N., Englert J. A., Nanoparticle delivery of microRNA-146a regulates mechanotransduction in lung macrophages and mitigates injury during mechanical ventilation. Nat. Commun. 12, 289 (2021). - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

