Vaccination with an mRNA-encoded membrane-bound HIV envelope trimer induces neutralizing antibodies in animal models
- PMID: 40737430
- PMCID: PMC12363394
- DOI: 10.1126/scitranslmed.adw0721
Vaccination with an mRNA-encoded membrane-bound HIV envelope trimer induces neutralizing antibodies in animal models
Abstract
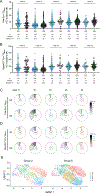
A protective vaccine against human immunodeficiency virus (HIV) will likely need to induce broadly neutralizing antibodies (bnAbs) that engage relatively conserved epitopes on the HIV envelope glycoprotein (Env) trimer. Nearly all vaccine strategies to induce bnAbs require the use of complex immunization regimens involving a series of different immunogens, most of which are Env trimers. Producing protein-based clinical material to evaluate such relatively complex regimens in humans presents major challenges in cost and time. Furthermore, immunization with HIV trimers as soluble proteins induces strong nonneutralizing responses to the trimer base, which is normally occluded on the virion. These base responses could potentially detract from the elicitation of nAbs and the eventual induction of bnAbs. mRNA vaccine platforms offer potential advantages over protein delivery for HIV vaccine development, including increased production speed, reduced cost, and the ability to deliver membrane-bound trimers that might facilitate improved immuno-focusing to nonbase epitopes. We report the design of mRNA-delivered soluble and membrane-bound forms of a stabilized native-like Env trimer (BG505 MD39.3); initial immunogenicity evaluation in rabbits that triggered clinical evaluation; and more comprehensive evaluation of B cell, T cell, and antibody responses in nonhuman primates. mRNA-encoded membrane-bound Env immunization elicited reduced off-target base-directed Env responses and stronger nAb responses compared with mRNA-encoded soluble Env. Overall, mRNA delivery of membrane-bound Env appears promising for enhancing B cell responses to subdominant epitopes and facilitating rapid translation to clinical testing, which should assist HIV vaccine development.
Conflict of interest statement
Figures




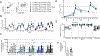
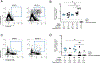
Update of
-
Vaccination with mRNA-encoded membrane-bound HIV Envelope trimer induces neutralizing antibodies in animal models.bioRxiv [Preprint]. 2025 Jan 25:2025.01.24.634423. doi: 10.1101/2025.01.24.634423. bioRxiv. 2025. Update in: Sci Transl Med. 2025 Jul 30;17(809):eadw0721. doi: 10.1126/scitranslmed.adw0721. PMID: 39896562 Free PMC article. Updated. Preprint.
References
-
- Steichen JM, Phung I, Salcedo E, Ozorowski G, Willis JR, Baboo S, Liguori A, Cottrell CA, Torres JL, Madden PJ, Ma KM, Sutton HJ, Lee JH, Kalyuzhniy O, Allen JD, Rodriguez OL, Adachi Y, Mullen TM, Georgeson E, Kubitz M, Burns A, Barman S, Mopuri R, Metz A, Altheide TK, Diedrich JK, Saha S, Shields K, Schultze SE, Smith ML, Schiffner T, Burton DR, Watson CT, Bosinger SE, Crispin M, Yates JR 3rd, Paulson JC, Ward AB, Sok D, Crotty S, Schief WR, Vaccine priming of rare HIV broadly neutralizing antibody precursors in nonhuman primates. Science 384, eadj8321 (2024); published online EpubMay 17 ( 10.1126/science.adj8321). - DOI - PMC - PubMed
-
- Caniels TG, Medina-Ramirez M, Zhang S, Kratochvil S, Xian Y, Koo JH, Derking R, Samsel J, van Schooten J, Pecetta S, Lamperti E, Yuan M, Carrasco MR, Del Moral Sanchez I, Allen JD, Bouhuijs JH, Yasmeen A, Ketas TJ, Snitselaar JL, Bijl TPL, Martin IC, Torres JL, Cupo A, Shirreff L, Rogers K, Mason RD, Roederer M, Greene KM, Gao H, Silva CM, Baken IJL, Tian M, Alt FW, Pulendran B, Seaman MS, Crispin M, van Gils MJ, Montefiori DC, McDermott AB, Villinger FJ, Koup RA, Moore JP, Klasse PJ, Ozorowski G, Batista FD, Wilson IA, Ward AB, Sanders RW, Germline-targeting HIV vaccination induces neutralizing antibodies to the CD4 binding site. Sci Immunol 9, eadk9550 (2024); published online EpubAug 30 ( 10.1126/sciimmunol.adk9550). - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

