Tumor-associated neutrophil precursors impair homologous DNA repair and promote sensitivity to PARP inhibition
- PMID: 40738898
- PMCID: PMC12310969
- DOI: 10.1038/s41467-025-61422-9
Tumor-associated neutrophil precursors impair homologous DNA repair and promote sensitivity to PARP inhibition
Abstract
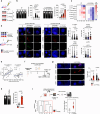
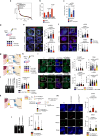
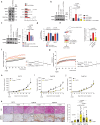
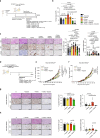
Tumor evolution is one of the major mechanisms responsible for acquiring therapy-resistant and more aggressive cancer clones. Whether the tumor microenvironment through immune-mediated mechanisms might promote the development of more aggressive cancer types is crucial for the identification of additional therapeutic opportunities. Here, we identify a subset of tumor-associated neutrophils, defined as tumor-associated neutrophil precursors (PreNeu). These PreNeu are enriched in highly proliferative hormone-dependent breast cancers and impair DNA repair capacity. Mechanistically, succinate secreted by tumor-associated PreNeu inhibits homologous recombination, promoting error-prone DNA repair through non-homologous end-joining regulated by PARP-1. Consequently, breast cancer cells acquire genomic instability promoting tumor editing and progression. Selective inhibition of these pathways induces increased tumor cell killing in vitro and in vivo. Tumor-associated PreNeu score correlates with copy number alterations in highly proliferative hormone-dependent tumors from breast cancer patients. Treatment with PARP-1 inhibitors counteract the pro-tumoral effect of these neutrophils and synergize with endocrine therapy.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare that Dr. Simon Barry is affiliated with IMED Oncology AstraZeneca, Li Ka Shing Center, Cambridge, United Kingdom and provided the αCXCR2 compound. The authors declare no competing interests.
Figures

References
-
- Falck, A. K., Ferno, M., Bendahl, P. O. & Ryden, L. St Gallen molecular subtypes in primary breast cancer and matched lymph node metastases–aspects on distribution and prognosis for patients with luminal A tumours: results from a prospective randomised trial. BMC Cancer13, 558 (2013). - PMC - PubMed
-
- DeSantis, C. E. et al. Breast cancer statistics, 2019. CA Cancer J. Clin.69, 438–451 (2019). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

