Candidate imprinting control regions in dog genome
- PMID: 40739170
- PMCID: PMC12309131
- DOI: 10.1186/s12864-025-11801-9
Candidate imprinting control regions in dog genome
Abstract
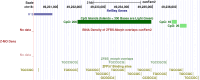
In mammals, genomic imprinting restricts the expression of a subset of genes from one of the two parental alleles. The process is regulated by imprinting control regions (ICRs) dispersed across autosomal chromosomal DNA. An unresolved question is how to discover candidate ICRs across the entire canine genome. Previously, bioinformatics analyses found a significant fraction of well-known ICRs in mouse, human, and Bos taurus. Analyses were based on finding the genomic positions of clusters of several CpG-rich motifs known as ZFBS-morph overlaps. These motifs are composite DNA elements. For this report, we performed similar studies to pinpoint candidate ICRs in the dog genome. A key feature of the bioinformatics approach is creating density plots to mark cluster positions as peaks. In genome-wide analyses, peaks in plots effectively discovered candidate ICRs along chromosomal DNA sequences of the Canis familiaris breed Boxer. With respect to Non-Dog RefSeq Genes, several candidate ICRs are in regions analogous to ICR positions in mouse DNA, in human DNA, or both. In the Boxer genome, examples include candidate ICRs for parent-of-origin-specific expression of the MEST isoform PEG1, INPP5F_V2, the PLAGL1 isoform ZAC1, IGF2R, PEG3, and GNAS loci. In mouse, imprinted genes in these loci play important roles in developmental and physiological processes.
Keywords: Canis familiaris; Allele-specific; Canine; CpG; DMR; Dog genome; Gene regulation; Genome-wide; ICR; Imprinted genes.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The research did not include human or animal subjects. Competing interests: The authors declare no competing interests.
Figures






Similar articles
-
Along the Bos taurus genome, uncover candidate imprinting control regions.BMC Genomics. 2022 Jun 28;23(1):478. doi: 10.1186/s12864-022-08694-3. BMC Genomics. 2022. PMID: 35764919 Free PMC article.
-
Discovering candidate imprinted genes and imprinting control regions in the human genome.BMC Genomics. 2020 May 31;21(1):378. doi: 10.1186/s12864-020-6688-8. BMC Genomics. 2020. PMID: 32475352 Free PMC article.
-
Effects of Developmental Lead and Phthalate Exposures on DNA Methylation in Adult Mouse Blood, Brain, and Liver: A Focus on Genomic Imprinting by Tissue and Sex.Environ Health Perspect. 2024 Jun;132(6):67003. doi: 10.1289/EHP14074. Epub 2024 Jun 4. Environ Health Perspect. 2024. PMID: 38833407 Free PMC article.
-
A systematic review and meta-analysis of DNA methylation levels and imprinting disorders in children conceived by IVF/ICSI compared with children conceived spontaneously.Hum Reprod Update. 2014 Nov-Dec;20(6):840-52. doi: 10.1093/humupd/dmu033. Epub 2014 Jun 24. Hum Reprod Update. 2014. PMID: 24961233
-
Mom genes and dad genes: genomic imprinting in the regulation of social behaviors.Epigenomics. 2025 Jun;17(8):555-573. doi: 10.1080/17501911.2025.2491294. Epub 2025 Apr 18. Epigenomics. 2025. PMID: 40249667 Review.
References
-
- Tucci V, Isles AR, Kelsey G, Ferguson-Smith AC, Erice Imprinting G. Genomic imprinting and physiological processes in mammals. Cell. 2019;176(5):952–65. - PubMed
-
- Proudhon C, Bourc’his D. [Evolution of genomic imprinting in mammals: what a zoo!]. Med Sci (Paris). 2010;26(5):497–503. - PubMed
-
- Radford EJ, Ferron SR, Ferguson-Smith AC. Genomic imprinting as an adaptative model of developmental plasticity. FEBS Lett. 2011;585(13):2059–66. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

