Identification of differentially expressed immune-related genes in patients with systemic lupus erythematosus and the development of a hub gene-based diagnostic model
- PMID: 40739239
- PMCID: PMC12308984
- DOI: 10.1186/s40001-025-02953-1
Identification of differentially expressed immune-related genes in patients with systemic lupus erythematosus and the development of a hub gene-based diagnostic model
Abstract
Background: Systemic lupus erythematosus (SLE) is an incurable autoimmune disease that affects body tissues, but it can be managed with medication. Although therapeutic strategies for SLE have advanced, the underlying molecular mechanisms driving disease pathogenesis remain incompletely understood.
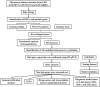
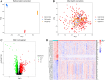
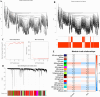
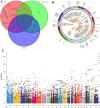
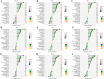
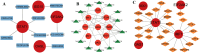
Methods: This study analyzed gene expression data from three GEO microarray datasets to explore immunity-related differentially expressed genes (DEGs) in SLE. Using WGCNA, we identified gene modules and integrated them with immune-related DEGs to find candidate hub genes, which were validated using RT-qPCR. We constructed a PPI network and performed gene enrichment analysis to identify nine hub genes through ROC curve analysis. We confirmed the link between these hub genes and immune cells, conducted GSEA, and predicted drugs, miRNAs, and transcription factors (TFs) targeting these genes. LASSO and ROC analyses validated a model using immunity-related DEGs.
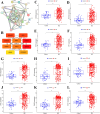
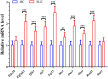
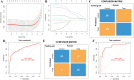
Results: The forty immune-related DEGs were identified from a total of 1590 DEGs, 452 WGCN module genes, and 1791 immune genes. Nine hub genes (MX1, OAS1, OASL, IRF7, RSAD2, EIF2AK2, ISG15, IFIH1, and STAT1) were highlighted using Cytoscape and ROC analysis, with an AUC greater than 0.7. RT-qPCR confirmed significant overexpression of all hub genes except STAT1 in SLE. ssGSEA and GSEA linked these genes to immune cell infiltration and pathways, including "cell cycle" and "RIG-I-like receptor signaling." A diagnostic model with three immune-related hub genes (MX1, IRF7, and EIF2AK2) demonstrated high accuracy (AUC > 0.8) in distinguishing SLE from healthy controls. Additionally, 9 target drugs, 14 target miRNAs, and 23 TFs were identified for these hub genes.
Conclusions: MX1, IRF7, and EIF2AK2 may serve as candidate biomarkers for SLE and warrant further investigation.
Keywords: Bioinformatics analysis; Diagnostic model; EIF2AK2; IRF7; Immune cell infiltration; MX1; Systemic lupus erythematosus.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: GEO is a public database containing ethically approved patient data. Users can freely download this data for research and publication purposes. This study, which utilizes open-source data from GEO, has no ethical concerns or conflicts of interest. Human protocols adhered to the Ethical Guidelines of the Declaration of Helsinki. The Jiangxi Provincial People's Hospital Ethics Committee (JXPPH-202310017) approved this study. All participants provided written informed consent, and all procedures followed relevant guidelines and regulations. Consent for publication: All authors know and approve the publication of this manuscript. Competing interests: The authors declare no competing interests.
Figures



References
-
- Siegel CH, Sammaritano LR. Systemic lupus erythematosus: A review. JAMA. 2024;331(17):1480–91. - PubMed
-
- Hoi A, Igel T, Mok CC, Arnaud L. Systemic lupus erythematosus. Lancet (London, England). 2024;403(10441):2326–38. - PubMed
-
- Karp DR. Complement and systemic lupus erythematosus. Curr Opin Rheumatol. 2005;17(5):538–42. - PubMed
-
- Kaul A, Gordon C, Crow MK, Touma Z, Urowitz MB, van Vollenhoven R, et al. Systemic lupus erythematosus. Nat Rev Dis Primers. 2016;2:16039. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

