PEX39 facilitates the peroxisomal import of PTS2-containing proteins
- PMID: 40739340
- PMCID: PMC12339391
- DOI: 10.1038/s41556-025-01711-z
PEX39 facilitates the peroxisomal import of PTS2-containing proteins
Abstract
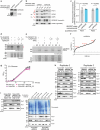
Peroxisomes are metabolic organelles essential for human health. Defects in peroxisomal biogenesis proteins (also known as peroxins (PEXs)) cause devastating disease. PEX7 binds proteins containing a type 2 peroxisomal targeting signal (PTS2) to enable their import from the cytosol into peroxisomes, although many aspects of this process remain enigmatic. Utilizing in vitro assays, yeast and human cells, we show that PEX39, a previously uncharacterized protein, is a cytosolic peroxin that facilitates the import of PTS2-containing proteins by binding PEX7 and stabilizing its interaction with cargo proteins containing a PTS2. PEX39 and PEX13, a peroxisomal membrane translocon protein, both possess an (R/K)PWE motif necessary for PEX7 binding. Handover of PEX7 from PEX39 to PEX13 via these motifs provides a new paradigm for peroxisomal protein import and biogenesis. Collectively, this work reveals how PEX39 and (R/K)PWE motifs facilitate the import of PTS2-containing proteins and advances our understanding of peroxisomal disease.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: R.J.D. is a founder and advisor at Atavistik Bio and serves on the scientific advisory boards of Agios Pharmaceuticals and Vida Ventures. The other authors declare no competing interests.
Figures

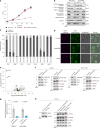
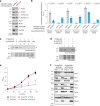







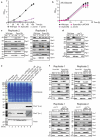
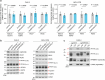





References
-
- Yifrach, E. et al. Defining the mammalian peroxisomal proteome. Subcell. Biochem.89, 47–66 (2018). - PubMed
-
- Waterham, H. R., Ferdinandusse, S. & Wanders, R. J. Human disorders of peroxisome metabolism and biogenesis. Biochim. Biophys. Acta1863, 922–933 (2016). - PubMed
-
- Francisco, T. et al. Protein transport into peroxisomes: knowns and unknowns. BioEssays39, 1700047 (2017). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

