Respiratory viral infections awaken metastatic breast cancer cells in lungs
- PMID: 40739350
- PMCID: PMC12422975
- DOI: 10.1038/s41586-025-09332-0
Respiratory viral infections awaken metastatic breast cancer cells in lungs
Abstract
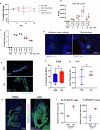
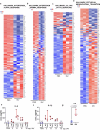
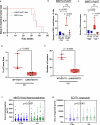
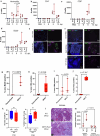
Breast cancer is the second most common cancer globally, with most deaths caused by metastatic disease, often following long periods of clinical dormancy1. Understanding the mechanisms that disrupt the quiescence of dormant disseminated cancer cells (DCCs) is crucial for addressing metastatic progression. Infections caused by respiratory viruses such as influenza and SARS-CoV-2 trigger both local and systemic inflammation2,3. Here we demonstrate, in mice, that influenza and SARS-CoV-2 infections lead to loss of the pro-dormancy phenotype in breast DCCs in the lung, causing DCC proliferation within days of infection and a massive expansion of carcinoma cells into metastatic lesions within two weeks. These phenotypic transitions and expansions are interleukin-6 dependent. We show that DCCs impair lung T cell activation and that CD4+ T cells sustain the pulmonary metastatic burden after the influenza infection by inhibiting CD8+ T cell activation and cytotoxicity. Crucially, these experimental findings align with human observational data. Analyses of cancer survivors from the UK Biobank (all cancers) and Flatiron Health (breast cancer) databases reveal that SARS-CoV-2 infection substantially increases the risk of cancer-related mortality and lung metastasis compared with uninfected cancer survivors. These discoveries underscore the huge impact of respiratory viral infections on metastatic cancer resurgence, offering new insights into the connection between infectious diseases and cancer metastasis.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: J.A.A.-G. is a co-founder, advisory board member and equity holder in HiberCell, a Mount Sinai spin-off that develops cancer recurrence-prevention therapies. He consults for HiberCell and Astrin Biosciences, serves as chief mission advisor for the Samuel Waxman Cancer Research Foundation and has ownership interest in patent number WO2019191115A1/ EP-3775171-B1. J.D. and M.R. are on the scientific advisory board for Mitotherapeutix. J.C.C. is a cofounder and chief scientific officer of OncoRx Insights. M.C.-H. holds shares in the O-SMOSE company and has no conflict of interest to disclose; consulting activities conducted by the company are independent of the present work. H.M. has consulted for Astra Zeneca relating to the use of monoclonal antibodies in the prevention and treatment of SARS-CoV-2 infection. All other authors declare no competing interests.
Figures











Update of
-
Respiratory viral infection promotes the awakening and outgrowth of dormant metastatic breast cancer cells in lungs.Res Sq [Preprint]. 2024 Apr 5:rs.3.rs-4210090. doi: 10.21203/rs.3.rs-4210090/v1. Res Sq. 2024. Update in: Nature. 2025 Sep;645(8080):496-506. doi: 10.1038/s41586-025-09332-0. PMID: 38645169 Free PMC article. Updated. Preprint.
References
-
- Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin.70, 7–30 (2020). - PubMed
-
- Soni, A. et al. Breast cancer subtypes predispose the site of distant metastases. Am. J. Clin. Pathol.143, 471–478 (2015). - PubMed
-
- Phan, T. G. & Croucher, P. I. The dormant cancer cell life cycle. Nat. Rev. Cancer20, 398–411 (2020). - PubMed
MeSH terms
Substances
Grants and funding
- R01 CA301643/CA/NCI NIH HHS/United States
- R01 AG078814/AG/NIA NIH HHS/United States
- R01 CA260909/CA/NCI NIH HHS/United States
- T32 GM141742/GM/NIGMS NIH HHS/United States
- P30 CA046934/CA/NCI NIH HHS/United States
- T32 AR079114/AR/NIAMS NIH HHS/United States
- R01 AI173305/AI/NIAID NIH HHS/United States
- P30 AG067988/AG/NIA NIH HHS/United States
- I01 BX004495/BX/BLRD VA/United States
- R01 AG081226/AG/NIA NIH HHS/United States
- R01 CA109182/CA/NCI NIH HHS/United States
- P30 CA013330/CA/NCI NIH HHS/United States
- R01 CA259635/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

