Immunoinformatic design of chimeric multiepitope vaccine for the prevention of human metapneumovirus (hMPV)
- PMID: 40739488
- PMCID: PMC12312241
- DOI: 10.1186/s12879-025-11339-x
Immunoinformatic design of chimeric multiepitope vaccine for the prevention of human metapneumovirus (hMPV)
Erratum in
-
Correction: Immunoinformatic design of chimeric multiepitope vaccine for the prevention of human metapneumovirus (hMPV).BMC Infect Dis. 2025 Sep 2;25(1):1093. doi: 10.1186/s12879-025-11493-2. BMC Infect Dis. 2025. PMID: 40898076 Free PMC article. No abstract available.
Abstract
Background: Human metapneumovirus (hMPV) is a significant etiological agent of acute respiratory infections in children and immunocompromised individuals. Despite its growing clinical impact, no approved vaccines or targeted antiviral therapies are currently available.
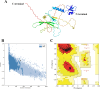
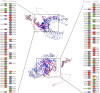
Methods: An immunoinformatic approach was employed to design a chimeric multi-epitope vaccine candidate against hMPV. Conserved and virulence-associated proteins were analyzed to predict highly antigenic B cell, cytotoxic T lymphocyte (CTL), and helper T lymphocyte (HTL) epitopes. The selected epitopes were screened for antigenicity, non-toxicity, non-allergenicity, and lack of homology to human proteins. The final construct included six B cell epitopes, six CTL epitopes, and two HTL epitopes, linked with appropriate adjuvants and Toll-like receptor (TLR) agonists. Structural modeling, molecular docking, and molecular dynamics simulations were conducted to evaluate the stability and receptor binding. Immunogenicity and expression potential were assessed through in silico immune simulation and codon optimization for expression in Escherichia coli.
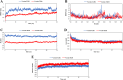
Results: All selected epitopes showed high antigenicity with no allergenic or toxicity. Structural validation indicated a stable vaccine construct with favorable physicochemical properties. Molecular docking analysis predicted a high binding affinity between the vaccine construct and TLR2/TLR4 receptors. Molecular dynamics (MD) simulations suggested that the docked complexes maintained stable interactions under simulated physiological conditions. In silico immune simulations predicted strong B- and T-cell responses following three doses. Codon adaptation analysis supported the high-level expression in E. coli.
Conclusion: The proposed multi-epitope vaccine demonstrates strong potential against hMPV, as supported by comprehensive computational analyses. Further experimental studies are required to validate its efficacy and safety.
Keywords: HMPV; Immunoinformatic; MD simulation; Molecular Docking; Multi-epitope vaccine.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures





References
-
- Turner P, XMGRACE. Version 5.1. 19. Center for coastal and Land-Margin research. Volume 2. Beaverton, OR: Oregon Graduate Institute of Science and Technology; 2005.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

