Pannexin-1 channels, extracellular ATP, and purinergic receptors are essential for CCR5/CXCR4 clustering and HIV entry
- PMID: 40740680
- PMCID: PMC12304881
- DOI: 10.1515/nipt-2025-0005
Pannexin-1 channels, extracellular ATP, and purinergic receptors are essential for CCR5/CXCR4 clustering and HIV entry
Abstract
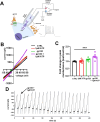
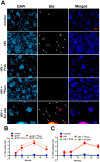
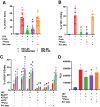
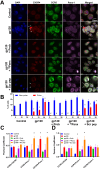
Objective: The Human Immunodeficiency Virus-1 (HIV) cell entry has been well characterized with the identification of CD4 as the main receptor and CXCR4 and CCR5 as co-receptors for the virus. However, how the virus uses the cell machinery for entry and infection is still a work-in-progress. Previously, we identified that the Pannexin-1 (Panx-1) channel, extracellular ATP, and purinergic receptors axis are essential for HIV entry and replication in macrophages, but the mechanisms were not fully explored.
Methods: Electrophysiology, ATP quantifications, confocal, HIV entry and replication experiments were used to determine the role of Panx-1 channels in HIV entry.
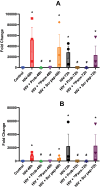
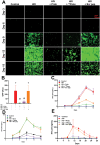
Results: Here, we identified that HIV or gp120 induces Panx-1 channel opening in association with ATP secretion, purinergic activation, and CCR5/CXCR4/actin clustering to enable HIV entry. Blocking Panx-1 channel opening, ATP secretion, or purinergic signaling prevented co-receptor clustering, HIV entry, and subsequent replication in multiple cell types.
Conclusion: We conclude that gp120 binding to the cell induces Panx-1 opening to promote the clustering of CCR5 or CXCR4 to the site of CD4-gp120 contact to aid viral entry.
Keywords: AIDS; HIV-1; cure; hemichannels; viral reservoirs.
© 2025 the author(s), published by De Gruyter, Berlin/Boston.
Conflict of interest statement
Conflict of interest: The authors have no financial conflicts of interest.
Figures

Similar articles
-
The role of Nef in the long-term persistence of the replication-competent HIV reservoir in South African women.J Virol. 2025 Jul 22;99(7):e0021725. doi: 10.1128/jvi.00217-25. Epub 2025 Jun 24. J Virol. 2025. PMID: 40552830 Free PMC article.
-
Rab11-FIP1C Is Dispensable for HIV-1 Replication in Primary CD4+ T Cells, but Its Role Is Cell Type Dependent in Immortalized Human T-Cell Lines.J Virol. 2022 Dec 14;96(23):e0087622. doi: 10.1128/jvi.00876-22. Epub 2022 Nov 10. J Virol. 2022. PMID: 36354340 Free PMC article.
-
Antiretrovirals for reducing the risk of mother-to-child transmission of HIV infection.Cochrane Database Syst Rev. 2011 Jul 6;(7):CD003510. doi: 10.1002/14651858.CD003510.pub3. Cochrane Database Syst Rev. 2011. PMID: 21735394
-
Patterns of inflammation and immune activation by coreceptor use in people living with HIV-1.Front Immunol. 2025 Jul 10;16:1632287. doi: 10.3389/fimmu.2025.1632287. eCollection 2025. Front Immunol. 2025. PMID: 40709192 Free PMC article.
-
Progressive resistive exercise interventions for adults living with HIV/AIDS.Cochrane Database Syst Rev. 2004 Oct 18;(4):CD004248. doi: 10.1002/14651858.CD004248.pub2. Cochrane Database Syst Rev. 2004. PMID: 15495092
References
LinkOut - more resources
Full Text Sources
Research Materials
