The Iris File Extension
- PMID: 40740735
- PMCID: PMC12309590
- DOI: 10.1016/j.jpi.2025.100461
The Iris File Extension
Abstract
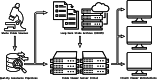
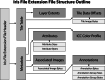
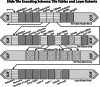
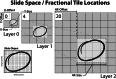
A modern digital pathology vendor-agnostic binary slide format specifically targeting the unmet need of efficient real-time transfer and display has not yet been established. The growing adoption of digital pathology only intensifies the need for an intermediary digital slide format that emphasizes performance for use between slide servers and image management software. The DICOM standard is a well-established format widely used for the long-term storage of both images and associated critical metadata. However, it was inherently designed for radiology rather than digital pathology, a discipline that imposes a unique set of performance requirements due to high-speed multi-pyramidal rendering within whole slide viewer applications. Here, we introduce the Iris file extension, a binary container specification explicitly designed for performance-oriented whole slide image (WSI) viewer systems. The Iris file extension specification is explicit and straightforward, adding modern compression support, a dynamic structure with fully optional metadata features, computationally trivial deep file validation, corruption recovery capabilities, and slide annotations. In addition to the file specification document, we provide source code to allow for (de)serialization and validation of a binary stream against the standard. We also provide corresponding binary builds with C++, Python, and JavaScript language support. Finally, we provide full encoder and decoder implementation source code, as well as binary builds (part of the separate Iris Codec Community module), with language bindings for C++ and Python, allowing for easy integration with existing WSI solutions. We provide the Iris File Extension specification openly to the community in the form of a Creative Commons Attribution-No Derivative 4.0 International license.
Keywords: DICOM; Digital pathology; File format; File specification; Iris; Performance digital pathology; Whole slide image.
© 2025 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this article.
Figures



References
-
- Landvater R.E., Balis U. Iris: a next generation digital pathology rendering engine. J Pathol Inform. 2025;16 doi: 10.1016/j.jpi.2024.100414. https://linkinghub.elsevier.com/retrieve/pii/S2153353924000531 URL. - DOI - PMC - PubMed
-
- Goode A., Gilbert B., Harkes J., Jukic D., Satyanarayanan M. Openslide: a vendor-neutral software foundation for digital pathology. J Pathol Inform. 2013;4:27. doi: 10.4103/2153-3539.119005. https://linkinghub.elsevier.com/retrieve/pii/S2153353922006484 URL. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

