Identification and validation of biomarkers, construction of diagnostic models, and investigation of immunological infiltration characteristics for idiopathic frozen shoulder
- PMID: 40740765
- PMCID: PMC12307173
- DOI: 10.3389/fimmu.2025.1559422
Identification and validation of biomarkers, construction of diagnostic models, and investigation of immunological infiltration characteristics for idiopathic frozen shoulder
Abstract
Background: Idiopathic frozen shoulder (FS) can lead to difficulties in daily activities and significantly impact the quality of life. Early diagnosis and treatment can help alleviate symptoms and restore shoulder function. Therefore, we aimed to explore the diagnostic biomarkers and potential mechanisms of FS from a transcriptomics perspective.
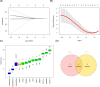
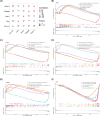
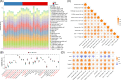
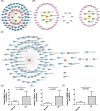
Methods: Total RNA was extracted from tissue samples of 15 FS and 11 controls. At the outset, we conducted differential expression analysis, weighted gene co-expression network analysis (WGCNA), and utilized the cytoHubba plugin, complemented by two machine learning algorithms, receiver operating characteristic (ROC) analysis, and expression level evaluation to identify biomarkers for FS. Subsequently, a nomogram was constructed based on the biomarkers. Additionally, we conducted enrichment and immune infiltration analyses to explore the mechanisms associated with these biomarkers. Finally, we confirmed the expression patterns of the biomarkers at the clinical level through reverse transcription-quantitative polymerase chain reaction (RT-qPCR).
Results: SNAI1, TWIST1, COL1A1, TUBB2B, and DCN were identified as biomarkers for FS. The nomogram constructed based on them had a good predictive value for the occurrence of FS. Except for DCN, the other four genes were upregulated in FS samples, and the expression of SNAI1, TWIST1, and TUBB2B was also observed to be significantly upregulated in RT-qPCR. Moreover, these genes played important roles in pathways such as "ECM receptor interaction" and "lysosome". We also found that the infiltration abundances of 11 types of immune cells were significantly upregulated in the FS samples, and they were positively correlated with each other. Our biomarkers showed strong correlations with these immune cells; DCN generally displayed a negative correlation, while the other four genes were generally positively correlated.
Conclusion: This study established a link between FS biomarkers that have strong diagnostic potential and specific immune responses, highlighting possible targets for diagnosing and treating FS.
Keywords: bioinformatics; frozen shoulder; immune infiltration; nomogram; transcriptomics.
Copyright © 2025 Jiang, Shen, Pang, Wu, Li, Gong, Jin and Zhu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

