The dysbiosis of gut microbiota and dysregulation of metabolites in IgA nephropathy and membranous nephropathy
- PMID: 40740937
- PMCID: PMC12307357
- DOI: 10.3389/fmed.2025.1618947
The dysbiosis of gut microbiota and dysregulation of metabolites in IgA nephropathy and membranous nephropathy
Abstract
Introduction: Immunoglobulin A nephropathy (IgAN) and membranous nephropathy (MN) are among the most common forms of primary glomerular diseases, with a rising global incidence. Despite their clinical importance, the underlying pathogenesis of these diseases and the development of reliable non-invasive diagnostic tools remain inadequately understood. Accumulating evidence suggests that gut microbiota and its associated metabolites may play a crucial role in the development of kidney diseases via the gut-kidney axis. However, comprehensive studies integrating both microbiome and metabolomic data in IgAN and MN are still limited.
Methods: In this study, we performed integrated metagenomic sequencing and untargeted metabolomic profiling to investigate alterations in gut microbial composition and systemic metabolic changes associated with IgAN and MN. Fecal samples were collected from 24 patients with IgAN, 20 patients with MN, and 17 healthy controls. Microbial diversity and composition were assessed using metagenomic analysis, while metabolic profiles were evaluated through untargeted LC -MS-based metabolomics. Multivariate statistical analyses and biomarker modeling were employed to identify discriminative features and evaluate diagnostic performance. Microbiota-metabolite correlation networks were constructed to explore potential mechanistic links.
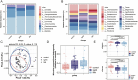
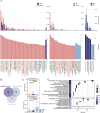
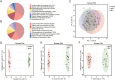
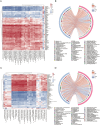
Results: Metagenomic analysis showed that both the IgAN and MN groups had significantly reduced α-diversity. Although β-diversity analysis did not reveal significant differences between the three groups, the IgAN and MN groups exhibited higher sample dispersion than the control group. Notably, both IgAN and MN patients showed a decrease in the abundance of certain specific microbial taxa. A total of 34 and 28 differentially abundant microbial species were identified in IgAN and MN, respectively, compared to healthy controls, with 16 taxa consistently downregulated in both disease groups. Notably, Streptococcus oralis was significantly enriched in the MN group, while [Clostridium] innocuum was markedly depleted. Metabolomic profiling identified 307 and 209 differentially abundant metabolites in IgAN and MN, respectively. Dipeptides (e.g., prolylleucine) were consistently upregulated, while the levels of certain short-chain fatty acids (SCFA) were reduced. Multivariate biomarker models demonstrated excellent diagnostic performance, achieving area under the curve (AUC) of 0.919 (IgAN vs. control), 0.897 (MN vs. control) and 0.912 (IgAN vs. MN), surpassing individual metabolite markers.
Discussion: Our findings highlight significant alterations in gut microbial composition and systemic metabolite profiles in both IgAN and MN patients compared to healthy individuals. The consistent reduction in microbial diversity and SCFA-producing taxa, along with characteristic changes in metabolic signatures, supports the involvement of the gut-kidney axis in disease pathogenesis. The diagnostic models developed in this study provide promising non-invasive biomarkers for distinguishing IgAN and MN with high accuracy. These results contribute novel insights into the microbe-metabolite interplay in glomerular diseases and offer potential targets for future diagnostic and therapeutic strategies.
Keywords: IgA nephropathy; biomarker; gut microbiota; membranous nephropathy; metabolomics.
Copyright © 2025 Zhang, Hu, Tan, Zhang, Chen, Gan, Chen, Zou, Wang, Pang, Fan and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Tao P, Huo J, Chen L. Bibliometric analysis of the relationship between gut microbiota and chronic kidney disease from 2001–2022. Integr Med Nephrol Androl. (2024) 11:17. doi: 10.1097/IMNA-D-23-00017 - DOI
-
- Zhao H, Zhao T, Li P. Gut microbiota-derived metabolites: a new perspective of traditional Chinese medicine against diabetic kidney disease. Integr Med Nephrol Androl. (2024) 11:24. doi: 10.1097/IMNA-D-23-00024 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

