Development and validation of machine learning-based diagnostic models using blood transcriptomics for early childhood diabetes prediction
- PMID: 40740938
- PMCID: PMC12308849
- DOI: 10.3389/fmed.2025.1636214
Development and validation of machine learning-based diagnostic models using blood transcriptomics for early childhood diabetes prediction
Abstract
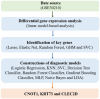
Background: Early identification of Type 1 Diabetes Mellitus (T1DM) in pediatric populations is crucial for implementing timely interventions and improving long-term outcomes. Peripheral blood transcriptomic analysis provides a minimally invasive approach for identifying predictive biomarkers prior to clinical manifestation. This study aimed to develop and validate machine learning algorithms utilizing transcriptomic signatures to predict T1DM onset in children up to 46 months before clinical diagnosis.
Methods: We analyzed 247 peripheral blood RNA-sequencing samples from pre-diabetic children and age-matched healthy controls. Differential gene expression analysis was performed using established bioinformatics pipelines to identify significantly dysregulated transcripts. Five feature selection methods (Lasso, Elastic Net, Random Forest, Support Vector Machine, and Gradient Boosting Machine) were employed to optimize gene sets. Nine machine learning algorithms (Decision Tree, Gradient Boosting Machine, K-Nearest Neighbors, Linear Discriminant Analysis, Logistic Regression, Multilayer Perceptron, Naive Bayes, Random Forest, and Support Vector Machine) were combined with selected features, generating 45 unique model combinations. Performance was evaluated using accuracy, precision, recall, and F1-score metrics. Model validation was conducted using quantitative polymerase chain reaction (qPCR) in an independent cohort of six children (three healthy, three diabetic).
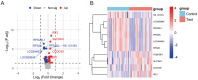
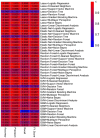
Results: Transcriptomic analysis revealed significant differential expression patterns between pre-diabetic and control groups. Four model combinations demonstrated superior predictive performance: Lasso+K-Nearest Neighbors, Elastic Net + K-Nearest Neighbors, Elastic Net + Random Forest, and Support Vector Machine+K-Nearest Neighbors. These models achieved high accuracy in predicting diabetes onset up to 46 months before clinical diagnosis. Both Elastic Net-based models achieved perfect classification performance in the validation cohort, demonstrating their potential as clinically viable diagnostic tools.
Conclusion: This study establishes the feasibility of integrating peripheral blood transcriptomic profiling with machine learning for early pediatric T1DM prediction. The identified transcriptomic signatures and validated predictive models provide a foundation for developing clinically translatable, non-invasive diagnostic tools. These findings support the implementation of precision medicine approaches for childhood diabetes prevention and warrant validation in larger, multi-center cohorts to assess generalizability and clinical utility.
Keywords: childhood diabetes; machine learning; pediatric biomarkers; peripheral blood; transcriptomic analysis.
Copyright © 2025 Huang, Ouyang, Xie, Zhuang, Gao, Liu and Guo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
LinkOut - more resources
Full Text Sources

