Multi-Omics Analysis Combined with Machine Learning Identified FABP4 in Smooth Muscle Cells as a Pathogenic Factor in Atherosclerosis
- PMID: 40740974
- PMCID: PMC12309571
- DOI: 10.2147/JIR.S519114
Multi-Omics Analysis Combined with Machine Learning Identified FABP4 in Smooth Muscle Cells as a Pathogenic Factor in Atherosclerosis
Abstract
Background: Atherosclerosis is the pathological basis of coronary heart disease, stroke, and peripheral arterial disease. Smooth muscle cells (SMCs) play a crucial role in atherosclerotic pathogenesis. However, effective drugs and therapy targeting SMCs for treating atherosclerosis are still lacking.
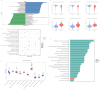
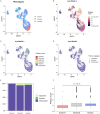
Methods: We utilized single-cell RNA sequencing (scRNA-seq) (GSE155512 and GSE159677) and array data (GSE43292 and GSE125771) to identify Scissor+ SMCs (SMCs positively associated with atherosclerosis) and Scissor- SMCs (SMCs negatively associated with atherosclerosis) by using Scissor package. We analyzed their functional changes, cell-cell communication, and differentiation potential. Machine learning techniques were employed to analyze the marker in SMCs of atherosclerosis. qRT-PCR was used to examine the expression of these genes in MOVAS stimulated by ox-LDL. Potential inhibitors of the identified proteins were predicted, and their binding sites were analyzed.
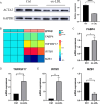
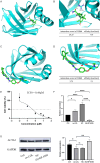
Results: We identified 475 Scissor+ SMCs and 1363 Scissor- SMCs. Functional enrichment analysis revealed that Scissor+ SMCs exhibited downregulation of Rho-related pathways, while pro-inflammatory pathways were upregulated. Cell-cell communication analysis indicated tighter interactions between SMCs and endothelial cells. Differential expression analysis identified 20 genes highly expressed in both scRNA-seq and array data. The LASSO regression, random forest, support vector machine and receiver operating characteristic curve suggested a strong correlation between fatty acid-binding protein 4 (FABP4) and atherosclerosis. The qRT-PCR results showed that FABP4 was highly expressed in MOVAS stimulated by ox-LDL. Drug prediction revealed that (S)-RP-6306 acted as an inhibitor, via forming a polar bond with Arg-126. In vitro experiments confirmed that (S)-RP-6306 significantly reduced the expression of FABP4.
Conclusion: Scissor+ SMCs differed significantly from Scissor- SMCs in cellular function, cell-cell communication, and differentiation potential. The high expression of FABP4 in this subgroup of SMCs presented a promising therapeutic target for atherosclerosis, with (S)-RP-6306 showing potential as a drug targeting FABP4.
Keywords: atherosclerosis; machine learning; molecular docking; smooth muscle cell.
© 2025 Wang et al.
Conflict of interest statement
The authors declare that they have no competing interests in this work.
Figures





References
LinkOut - more resources
Full Text Sources
Research Materials

