Development and validation of deep learning- and ensemble learning-based biological ages in the NHANES study
- PMID: 40741049
- PMCID: PMC12307447
- DOI: 10.3389/fnagi.2025.1532884
Development and validation of deep learning- and ensemble learning-based biological ages in the NHANES study
Abstract
Introduction: Conventional machine learning (ML) approaches for constructing biological age (BA) have predominantly relied on blood-based markers, limiting their scope. This study aims to develop and validate novel ML-based BA models using a comprehensive set of clinical, behavioral, and socioeconomic factors and evaluate their predictive performance for mortality.
Methods: We analyzed data from 24,985 participants in the National Health and Nutrition Examination Survey (NHANES) from 1999 to 2010, with follow-up extending to 31 December 2019, or until death or loss to follow-up. Thirty features, including blood and urine biochemistry, physical examination data, behavioral traits, and socioeconomic factors, were selected using the Least Absolute Shrinkage and Selection Operator (LASSO). These features were utilized to train deep neural networks (DNN) and ensemble learning models, specifically the Deep Biological Age (DBA) and Ensemble Biological Age (EnBA), with chronological age (CA) as the reference label. Model performance was assessed using mean absolute error (MAE), while interpretability was explored using Shapley Additive exPlanation (SHAP). Predictive accuracy of DBA and EnBA for mortality was compared with Phenotypic Age (PhenoAge) using the area under the curve (AUC) derived from Cox proportional hazards models and hazard ratios (HR), adjusted for demographics and lifestyle factors. Sensitivity analyses were performed to ensure robustness.
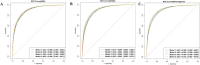
Results: DBA and EnBA accurately predicted actual age (MAE = 2.98 and 3.58 years, respectively) and demonstrated strong predictive capability for all-cause mortality, with AUCs of 0.896 (95% CI: 0.891-0.898) for DBA and 0.889 (95% CI: 0.884-0.894) for EnBA. Higher DBA and EnBA accelerations were significantly associated with increased mortality risk (HR = 1.059 and 1.039, respectively). SHAP analysis highlighted prescription medication usage, hepatitis B surface antibody status, and vigorous physical activity as the most influential features contributing to DBA predictions. Furthermore, BA acceleration was linked to elevated risk of death from specific chronic conditions, including cardiovascular and cerebrovascular diseases and cancer.
Discussion: Our study successfully developed and validated two ML-based BA models capable of accurately predicting both all-cause and cause-specific mortality. These findings suggest that the DBA and EnBA models hold promise for early identification of high-risk individuals, potentially facilitating timely preventive interventions and improving population health outcomes.
Keywords: aging; biological age; deep learning; deep neural networks; machine learning.
Copyright © 2025 Huang, Yang, Wang, Abula, Dong and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Cabral D., Bigliassi M., Cattaneo G., Rundek T., Pascual-Leone A., Cahalin L., et al. (2022). Exploring the interplay between mechanisms of neuroplasticity and cardiovascular health in aging adults: A multiple linear regression analysis study. Auton Neurosci. 242:103023. 10.1016/j.autneu.2022.103023 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

