Selecting differential splicing methods: Practical considerations for short-read RNA sequencing
- PMID: 40741371
- PMCID: PMC12308171
- DOI: 10.12688/f1000research.155223.2
Selecting differential splicing methods: Practical considerations for short-read RNA sequencing
Abstract
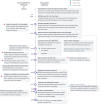
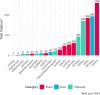
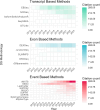
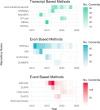
Alternative splicing is crucial in gene regulation, with significant implications in clinical settings and biotechnology. This review article compiles bioinformatics short-read RNA-seq tools for investigating differential splicing; offering a detailed examination of their statistical methods, case applications, and benefits. A total of 22 tools are categorised by their statistical family (parametric, non-parametric, and probabilistic) and level of analysis (transcript, exon, and event). The central challenges in quantifying alternative splicing include correct splice site identification and accurate isoform deconvolution of transcripts. Benchmarking studies show no consensus on tool performance, revealing considerable variability across different scenarios. Tools with high citation frequency and continued developer maintenance, such as DEXSeq and rMATS, are recommended for prospective researchers. To aid in tool selection, a guide schematic is proposed based on variations in data input and the required level of analysis. Emerging long-read RNA sequencing technologies are discussed as a complement to short-read methods, promising reduced deconvolution needs and further innovation.
Keywords: Alternative Splicing; Bioinformatics; Differential Expression; RNASeq; Transcriptomics.
Copyright: © 2025 Draper BJ et al.
Conflict of interest statement
No competing interests were disclosed.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

