Phenotypic Divergence of JAG1- and NOTCH2-Associated Alagille Syndrome & Disease-Specific NOTCH2 Variant Classification Guidelines
- PMID: 40742203
- PMCID: PMC12312628
- DOI: 10.1111/liv.70251
Phenotypic Divergence of JAG1- and NOTCH2-Associated Alagille Syndrome & Disease-Specific NOTCH2 Variant Classification Guidelines
Abstract
Background & aims: Alagille syndrome (ALGS) is a rare, autosomal dominant disorder with high phenotypic heterogeneity. Disease-causing variants are primarily identified in Jagged1 (JAG1), with fewer reported in NOTCH2. JAG1 variants cause disease through a mechanism of haploinsufficiency, but the mechanism for NOTCH2 variants is not completely understood, making classification of variants more challenging. Using a large, international patient cohort acquired through the Global ALagille Alliance (GALA) study, we sought to improve classification of NOTCH2 variants and study phenotypic differences between NOTCH2- and JAG1-related disease.
Methods: Clinical and molecular data from 952 individuals with ALGS in GALA were analysed and disease features compared between those with JAG1 (n = 902) and NOTCH2 (n = 34) variants. Previously reported and newly identified NOTCH2 variants were reinterpreted based on disease-specific modifications to the American College of Medical Genetics and Genomics (ACMG) guidelines. The Kaplan-Meier method was utilised to assess native liver survival (NLS) and overall survival (OS) and gene comparisons were made with the log-rank test.
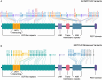
Results: Thirty NOTCH2 variants, including 18 novel variants, were identified and classified in our GALA cohort. Phenotypic analyses revealed a significantly lower incidence of characteristic facies, posterior embryotoxon, cardiac involvement and butterfly vertebrae in individuals with NOTCH2 variants compared to those with JAG1 variants (p < 0.001). No differences were identified in NLS or OS. Review of 61 previously reported NOTCH2 variants resulted in the re-classification of 19 likely pathogenic or pathogenic to VOUS (31.1%) with less than half retaining their originally published classification (34.4%; n = 21).
Conclusions: We report on a large global study on NOTCH2 genetics and phenotype, which increases the number of reported NOTCH2 variants by 30%. All variants were reclassified using current guidelines, and comparison of the JAG1 and NOTCH2 cohorts demonstrates clear phenotypic divergence between these groups. These data suggest that reliance on classical clinical phenotyping may miss patients with NOTCH2-related disease and supports an inclusive approach to genetic testing.
Keywords: Alagille syndrome; NOTCH2; cholestasis; genetics.
© 2025 The Author(s). Liver International published by John Wiley & Sons Ltd.
Conflict of interest statement
Shannon M. Vandriel [consults for Mirum Pharmaceuticals Inc.], Li‐Ting Li, Huiyu She, David A. Piccoli, Irena Jankowska, Piotr Czubkowski, Dorota Gliwicz‐Miedzińska, Emanuele Nicastro, Dominique Debray, Étienne M. Sokal, Tanguy Demaret, Rima L. Fawaz, Silvia Nastasio, Kyung Mo Kim, Seak Hee Oh, Catherine Larson‐Nath [nothing to disclose], Sahana Shankar, Shikha S. Sundaram, Alexander Chaidez, Pinar Bulut, Pier Luigi Calvo, Mureo Kasahara, Niviann Blondet, Eberhard Lurz, Anna‐Maria Kavallar, Emmanuel M. Gonzales, Jérôme Bouligand, Jeffrey A. Feinstein, Susan M. Siew, Michael O. Stormon, Rene Romero, M. Kyle Jensen, Catalina Jaramillo, James E. Squires, Sarah M. Bedoyan, Jane Hartley, Way Seah Lee, Chatmanee Lertudomphonwanit, Henry C. Lin, Yael Mozer‐Glassberg, Amin J. Roberts, Helen M. Evans, Gabriella Nebbia, Pamela L. Valentino, Jesus Quintero Bernabeu, Cigdem Arikan, María Legarda Tamara, Cristina Molera Busoms, Thomas Damgaard Sandahl, Andréanne N. Zizzo, Aglaia Zellos, Ruben E. Quiros‐Tejeira, Ermelinda Santos‐Silva, Jernej Brecelj, Maria Camila Sanchez, Maria Lorena Cavalieri, Christos Tzivinikos, Sabina Wiecek, John Eshun, Zerrin Önal, Cristina Gonçalves, Jennifer Garcia, Seema Alam, Carolina Jimenez‐Rivera, Luis Bujanda [nothing to disclose], Jian‐She Wang [Consultant for Ipsen Biopharmaceuticals Inc., Mirum Pharmaceuticals Inc., and Qing Bile Therapeutics Inc.], Kathleen M. Loomes [Consultant for Mirum Pharmaceuticals Inc., Ipsen Biopharmaceuticals Inc., and Travere Therapeutics Inc.], Lorenzo D'Antiga [Consultant for Mirum Pharmaceuticals Inc., Ipsen Biopharmaceuticals Inc., Alexion, Astra Zeneca, Vivet Therapeutics, Spark, Genespire, Tome, Advanza Pharma], Florence Lacaille [Consultant for Alexion], Björn Fischler [Consultant for Ipsen Biopharmaceuticals Inc.], Henrik Arnell [consultant for Ipsen Biopharmaceuticals Inc., Baxter, Mirum Pharmaceuticals Inc.], Winita Hardikar [Consultant for Ipsen Biopharmaceuticals Inc., Advanz Pharma], Emmanuel Jacquemin [Consultant for CTRS, Theravia and Vivet Therapeutics, France], Noelle H. Ebel [Consultant for Mirum Pharmaceuticals Inc.], Saul J. Karpen [Consultant for Ipsen Biopharmaceuticals Inc., Intercept, Mirum Pharmaceuticals Inc. and Vertex], Deirdre A. Kelly [European Advisory Board for Mirum Pharmaceuticals Inc.; Advisory Board for Intercept & Advanz Pharma], Henkjan J. Verkade [Consultant for Ipsen Biopharmaceuticals Inc., Intercept, Mirum Pharmaceuticals Inc., Orphalan and Vertex], Ryan T. Fischer [Speaker and Consultant for Ipsen Biopharmaceuticals Inc. and Mirum Pharmaceuticals Inc.], Nathalie Rock [Consultant for Mirum Pharmaceuticals Inc.], Wikrom Karnsakul [Consultant for Mirum Pharmaceuticals Inc., Ipsen Biopharmaceuticals Inc., and Travere Therapeutics Inc.], Victorien M. Wolters [Speaker for Mirum Pharmaceuticals Inc.], Amal A. Aqul [Consultant for Mirum Pharmaceuticals Inc., Ipsen Biopharmaceuticals Inc., and Sarepta], Giuseppe Indolfi [Consultant for Mirum Pharmaceuticals Inc.], Kathleen B. Schwarz [Research grant Ipsen Biopharmaceuticals Inc. and Consultant, Mirum Pharmaceuticals Inc], Nanda Kerkar [Advisory Board for Ipsen Biopharmaceuticals Inc. and Mirum Pharmaceuticals Inc.], Quais Mujawar [Advisory Board for Alexion Pharma], Richard J. Thompson [Consultant for Shire, Ipsen Biopharmaceuticals Inc., Mirum Pharmaceuticals Inc., Horizon Pharmaceuticals, Sana Biotechnology, GenerationBio, Retrophin and Qing Bile Therapeutics Inc.], Bettina E. Hansen [Consultant for Mirum Pharmaceuticals Inc., Ipsen Biopharmaceuticals Inc., Chemomab, Calliditas, Intercept, Cyma Bay, unrestricted grants from Cyma bay, Intercept, Mirum Pharmaceuticals Inc., and Ipsen Biopharmaceuticals Inc.], Nancy B. Spinner [Consultant for Mirum Pharmaceuticals Inc., and Travere Therapeutics Inc.], Melissa A. Gilbert [Consultant for Travere Therapeutics Inc.], Binita M. Kamath [Consultant for Mirum Pharmaceuticals Inc., Ipsen Biopharmaceuticals Inc., Third Rock Ventures and Audentes Therapeutics, unrestricted educational grants from Mirum Pharmaceuticals Inc., and Ipsen Biopharmaceuticals Inc.].
Figures

References
-
- Karpen S. J., Kamath B. M., Alexander J. J., et al., “Use of a Comprehensive 66‐Gene Cholestasis Sequencing Panel in 2171 Cholestatic Infants, Children, and Young Adults,” Journal of Pediatric Gastroenterology and Nutrition 72 (2021): 654–660. - PubMed
-
- Alagille D., Odievre M., Gautier M., and Dommergues J. P., “Hepatic Ductular Hypoplasia Associated With Characteristic Facies, Vertebral Malformations, Retarded Physical, Mental, and Sexual Development, and Cardiac Murmur,” Journal of Pediatrics 86 (1975): 63–71. - PubMed
-
- Emerick K. M., Rand E. B., Goldmuntz E., Krantz I. D., Spinner N. B., and Piccoli D. A., “Features of Alagille Syndrome in 92 Patients: Frequency and Relation to Prognosis,” Hepatology 29 (1999): 822–829. - PubMed
-
- Spinner N. B., Colliton R. P., Crosnier C., Krantz I. D., Hadchouel M., and Meunier‐Rotival M., “Jagged1 Mutations in Alagille Syndrome,” Human Mutation 17 (2001): 18–33. - PubMed
-
- Crosnier C., Lykavieris P., Meunier‐Rotival M., and Hadchouel M., “Alagille Syndrome. The Widening Spectrum of Arteriohepatic Dysplasia,” Clinics in Liver Disease 4 (2000): 765–778. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

