Elevated Transglutaminase-2 in SOX10-Deficient Melanoma Promotes Tumor Onset and Decreases Intratumoral CD4+ T Cells
- PMID: 40742313
- PMCID: PMC12398951
- DOI: 10.1158/0008-5472.CAN-24-3267
Elevated Transglutaminase-2 in SOX10-Deficient Melanoma Promotes Tumor Onset and Decreases Intratumoral CD4+ T Cells
Abstract
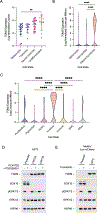
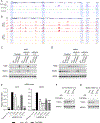
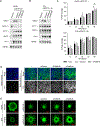
Melanoma heterogeneity contributes to therapy resistance and immune evasion. The loss of SOX10, a neural crest lineage-specific transcription factor, leads to phenotypic switching from a proliferative cell state to an invasive, drug-tolerant cell state. SOX10-deficient cells are able to persist during immunotherapy treatment, highlighting the need to characterize the factors that regulate immune evasion downstream of SOX10 loss. In this study, we found that SOX10-deficient melanoma cell lines and patient samples express elevated levels of TGM2, a transglutaminase family member. TGM2 upregulation in SOX10 knockout cells was reversed by inhibition of epigenetic reader BET proteins. Knockdown of TGM2 did not affect the SOX10-deficient invasive cell state; however, overexpression of TGM2 in syngeneic melanomas promoted tumor onset in immunocompetent mice, but not in immunodeficient mice, suggesting an immune-mediated effect. TGM2 overexpression in melanoma was associated with decreased intratumoral CD4+ T cells, and depletion of CD4+ T cells abolished the tumor-promoting effect of TGM2. These data indicate that TGM2 is negatively regulated by SOX10 in melanoma and can promote an immunosuppressive tumor microenvironment.
Significance: The transglutaminase TGM2 is negatively associated with the neural crest lineage-specific transcription factor SOX10 and is an immunomodulatory protein in cutaneous melanoma.
©2025 American Association for Cancer Research.
Conflict of interest statement
Figures




References
-
- Rambow F, Rogiers A, Marin-Bejar O, Aibar S, Femel J, Dewaele M et al. , Toward Minimal Residual Disease-Directed Therapy in Melanoma. Cell. 174, 843–855.e819 (2018). - PubMed
-
- Wouters J, Kalender-Atak Z, Minnoye L, Spanier KI, De Waegeneer M, Bravo González-Blas C et al. , Robust gene expression programs underlie recurrent cell states and phenotype switching in melanoma. Nat Cell Biol. 22, 986–998 (2020). - PubMed
MeSH terms
Substances
Grants and funding
- P30 CA016672/CA/NCI NIH HHS/United States
- T32 GM100836/GM/NIGMS NIH HHS/United States
- R01 CA196278/CA/NCI NIH HHS/United States
- R01 CA182635/CA/NCI NIH HHS/United States
- T32 CA236736/CA/NCI NIH HHS/United States
- P30 CA056036/CA/NCI NIH HHS/United States
- Melanoma Research Foundation (MRF)
- R01 CA182635/CA/NCI NIH HHS/United States
- R01 CA196278/CA/NCI NIH HHS/United States
- T32 CA236736/CA/NCI NIH HHS/United States
- T32 GM100836/GM/NIGMS NIH HHS/United States
- Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (AMRF)
- HT9425-23-MRP-MASA-ME230214/U.S. Department of Defense (DOD)
- W. W. Smith Charitable Trust (W. W. Smith Foundation)
- P30 CA056036/CA/NCI NIH HHS/United States
- P30 CA16672/National Institutes of Health (NIH)
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

