Novel Method for Predicting Lp(a) From Genomic Testing Identifies ASCVD Risk Across a Diverse Cohort
- PMID: 40742365
- PMCID: PMC12665494
- DOI: 10.1016/j.jacbts.2025.04.012
Novel Method for Predicting Lp(a) From Genomic Testing Identifies ASCVD Risk Across a Diverse Cohort
Abstract
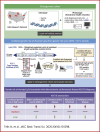
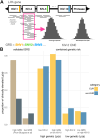
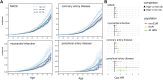
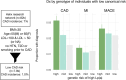
Lipoprotein(a) (Lp[a]) is a genetic and often unmeasured contributor to atherosclerotic cardiovascular disease (ASCVD) risk. In this study, Lp(a) was estimated from exome data by quantifying Kringle IV subtype 2 repeats alongside a single-nucleotide variant-based genetic risk score. This method was applied to a diverse cohort (N = 76,147) from the Helix Research Network. The method better identified individuals with high Lp(a) levels, especially among individuals not genetically similar to Europeans. High genetic risk for high Lp(a) level was correlated with earlier and more frequent ASCVD diagnoses. Those with high genetic Lp(a) were more likely to have ASCVD without traditional risk factors present.
Keywords: cardiovascular disease; clinical genetics; genomics; lipoprotein(a).
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Funding Support and Author Disclosures Funding was provided to the Healthy Nevada Project by the Renown Institute for Health Innovation and the Renown Health Foundation. The Healthy Nevada Project receives funding from Gilead Sciences, outside the scope of this research. Funding was provided to the myGenetics program by HealthPartners. Drs Barrett, Cirulli, Bolze, Telis, Lee, Luo, and Washington and Mr Khuder are employees of Helix. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Figures
References
-
- Marcovina S.M., Koschinsky M.L. Lipoprotein(a) as a risk factor for coronary artery disease. Am J Cardiol. 1998;82:57U–66U. - PubMed
-
- Tsimikas S., Marcovina S.M. Ancestry, lipoprotein(a), and cardiovascular risk thresholds: JACC review topic of the week. J Am Coll Cardiol. 2022;80:934–946. - PubMed
-
- Kraft H.G., Lingenhel A., Pang R.W., et al. Frequency distributions of apolipoprotein(a) kringle IV repeat alleles and their effects on lipoprotein(a) levels in Caucasian, Asian, and African populations: the distribution of null alleles is non-random. Eur J Hum Genet. 1996;4:74–87. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

