G-quadruplexes as potential traps for superenhancer marker BRD4: ligand-sensitive binding and co-separation in vitro
- PMID: 40744496
- PMCID: PMC12311796
- DOI: 10.1093/nar/gkaf726
G-quadruplexes as potential traps for superenhancer marker BRD4: ligand-sensitive binding and co-separation in vitro
Abstract
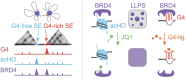
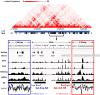
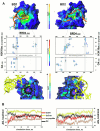
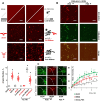
G-quadruplexes (G4s) are prevalent at promoters and superenhancers (SEs), exclude nucleosomes, and recruit transcription factors. This study sought to determine whether the nucleosome exclusion affects the recruitment of the SE marker BRD4, which typically binds to acetylated histones and facilitates SE-promoter contacts via the phase separation-dependent mechanism. Analyses of the available whole-genome data revealed that SEs with the highest G4 density were depleted of nucleosomes but not of BRD4. This led us to test the possibility of histone-independent BRD4 maintenance at G4-rich SEs. A typical SE G4 destabilized a nearby nucleosome in vitro and, unlike B-DNA, bound weakly to BRD4 bromodomains. Similar to an acetylated nucleosome, the G4 promoted phase separation in BRD4 solutions. This effect was not altered by the histone competitor JQ1. However, it was attenuated by two known G4 ligands, suggesting that they could disrupt SE-promoter communication in cells. Consistently, these ligands downregulated several genes regulated by G4-rich SE-contacting promoters more efficiently than they did SE-independent genes. Our findings underscore the significance of G4-rich SEs as transcriptional regulators and provide new insights into their organization.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

