A diverse landscape of FGFR alterations and co-mutations suggests potential therapeutic strategies in pediatric low-grade gliomas
- PMID: 40744913
- PMCID: PMC12314019
- DOI: 10.1038/s41467-025-61820-z
A diverse landscape of FGFR alterations and co-mutations suggests potential therapeutic strategies in pediatric low-grade gliomas
Erratum in
-
Author Correction: A diverse landscape of FGFR alterations and co-mutations suggests potential therapeutic strategies in pediatric low-grade gliomas.Nat Commun. 2025 Oct 9;16(1):8970. doi: 10.1038/s41467-025-64919-5. Nat Commun. 2025. PMID: 41068145 Free PMC article. No abstract available.
Abstract
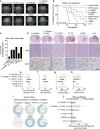
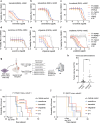
Oncogenic alterations in fibroblast growth factor receptor (FGFR)-family proteins occur across cancers, including pediatric gliomas. Our genomic analysis of 11,635 gliomas across ages finds that 5.3% of all gliomas harbor FGFR alterations, with an incidence of almost 9% in pediatric gliomas. Alterations in FGFR proteins are differentially enriched by age, tumor grade, and histology, with FGFR1 alterations associated with glioneuronal histologies. Leveraging isogenic systems, we confirm FGFR1 alterations to induce downstream Mitogen Activated Protein Kinase (MAPK) and mTOR signaling pathways, drive gliomagenesis, activate neuronal transcriptional programs and exhibit sensitivity to MAPK pathway and pan-FGFR inhibitors. Finally, we perform a retrospective analysis of clinical responses in children diagnosed with FGFR-altered gliomas and find that treatment with currently available inhibitors is largely associated with stability of disease. This study provides key insights into the biology of FGFR1-altered gliomas, therapeutic strategies to target them and associated challenges that still need to be overcome.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: P.B. serves on paid advisory boards for DayOne Biopharmaceuticals and has served on a paid advisory board for QED Therapeutics. Her lab has received grant funding from Novartis Institute of Biomedical Research. SHR has employment at Labcorp Oncology. C.M.V.T. is on advisory boards for Alexion, Bayer, Novartis, and Roche and has received travel support from Eli Lilly. J.W. is a consultant and equity holder for Soltego. F.S. and D.T.W.J. are co-founders and shareholders of Heidelberg Epignostix GmbH. O.W. is advisory board member or has received research grants from Janssen, Day One Biopharmaceuticals, and Novartis. K.L.L. disclosures: Equity: Travera Inc.; Consulting- Travera Inc., B.M.S., Servier, L.E.K., Integragen, Blaze Bioscience; Research. Research Funding to DFCI: B.M.S. K.M., M.M., L.A.A. are employees of Foundation Medicine, Inc. and stockholders of Roche Holdings AG. E.S.F. is a founder, scientific advisory board (SAB) member, and equity holder of Civetta Therapeutics, Proximity Therapeutics, Neomorph, Inc. (also board of directors), Stelexis Biosciences, Inc., Anvia Therapeutics, Inc., and CPD4, Inc. (also board of directors). He is an equity holder and SAB member for Avilar Therapeutics, Photys Therapeutics, and Ajax Therapeutics, and an equity holder in Lighthorse Therapeutics. E.S.F. is a consultant to Novartis, EcoR1 capital, Odyssey, and Deerfield. The Fischer lab receives or has received research funding from Deerfield, Novartis, Ajax, Interline, Bayer, and Astellas. The Eck lab has received sponsored research support from Novartis, and MJE is a consultant for Syndax Pharmaceuticals. The remaining authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

