PRMT1-mediated methylation of UBE2m promoting calcium oxalate crystal-induced kidney injury by inhibiting fatty acid metabolism
- PMID: 40744915
- PMCID: PMC12313907
- DOI: 10.1038/s41419-025-07888-3
PRMT1-mediated methylation of UBE2m promoting calcium oxalate crystal-induced kidney injury by inhibiting fatty acid metabolism
Abstract
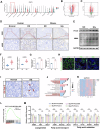
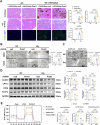
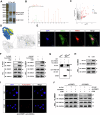
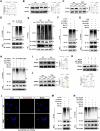
Calcium oxalate (CaOx) is the most common type of kidney stone, and its crystal deposition can induce oxidative stress, inflammatory responses, and cell death. This further aggravates kidney structural and functional damage, which in turn, promotes kidney stone recurrence, forming a vicious cycle of repeated stone formation and renal injury. Therefore, identifying precise and effective therapeutic targets is crucial to prevent the damage and inflammation caused by kidney stones. Protein arginine methyltransferase 1 (PRMT1) is a well-known epigenetic regulatory enzyme involved in renal metabolic reprogramming. However, the role of PRMT1-mediated arginine methylation in kidney stone-induced renal injury remains unclear. In this study, mice with specific deletion or overexpression of PRMT1 in tubular epithelial cells were developed, and a CaOx crystal-induced kidney injury mouse model was established. Single-cell RNA-sequencing, metabolomic, proteomic, and transcriptomic analyses, together with immunoprecipitation, mass spectrometry, GST-pulldown assays, oxygen consumption rate assays, and other methods, were used to reveal the mechanism of PRMT1 in renal injury caused by CaOx crystals. Specifically, PRMT1 enhanced the protein function of UBE2m through arginine methylation at R169, and increased the neddylation level and protein stability of NEDD4, thereby inducing PPARγ ubiquitination. Increased PPARγ degradation inhibited downstream fatty acid metabolism, leading to renal lipid accumulation, disrupted energy metabolism, and impaired kidney function. These findings provide a novel potential therapeutic target for CaOx kidney stones.
© 2025. The Author(s).
Conflict of interest statement
COMPETING INTERESTS: The authors declare no competing interests. Ethics approval and consent to participate: Samples were obtained with informed consent, and all protocols were performed in accordance with the Declaration of Helsinki and were approved by the review board of Renmin Hospital of Wuhan University (WDRY2021-KS047). Informed consent was obtained from patients. Animal experiments were approved by the Animal Care and Use Committee of Renmin Hospital of Wuhan University (WDRM 20200604), which complied with the Guide for the Care and Use of Laboratory Animals (NIH Publication No. 85-23). All methods were performed in accordance with the relevant guidelines and regulations.
Figures



Similar articles
-
Management of urinary stones by experts in stone disease (ESD 2025).Arch Ital Urol Androl. 2025 Jun 30;97(2):14085. doi: 10.4081/aiua.2025.14085. Epub 2025 Jun 30. Arch Ital Urol Androl. 2025. PMID: 40583613 Review.
-
Targeting PRMT1-mediated methylation of TAF15 to protect against myocardial infarction by inhibiting ferroptosis via the GPX4/NRF2 pathway.Clin Epigenetics. 2025 Jul 22;17(1):129. doi: 10.1186/s13148-025-01935-8. Clin Epigenetics. 2025. PMID: 40696470 Free PMC article.
-
ACOT4 and ACOT6 Activate Akt-mTOR Pathway and Inhibit Calcium Oxalate-Induced Renal Tubular Cell Injury.Kidney Blood Press Res. 2025;50(1):533-544. doi: 10.1159/000546897. Epub 2025 Jun 22. Kidney Blood Press Res. 2025. PMID: 40544826
-
Gut microbiota-derived indole-3-acetic acid ameliorates calcium oxalate renal stone formation via AHR/NF‑κB axis.Urolithiasis. 2025 Jul 2;53(1):134. doi: 10.1007/s00240-025-01779-0. Urolithiasis. 2025. PMID: 40601009 Free PMC article.
-
Citrate salts for preventing and treating calcium containing kidney stones in adults.Cochrane Database Syst Rev. 2015 Oct 6;2015(10):CD010057. doi: 10.1002/14651858.CD010057.pub2. Cochrane Database Syst Rev. 2015. PMID: 26439475 Free PMC article.
References
-
- Khan SR, Pearle MS, Robertson WG, Gambaro G, Canales BK, Doizi S, et al. Kidney stones. Nat Rev Dis Prim. 2017;3:17001. 10.1038/nrdp.2017.1. - PubMed
-
- Capolongo G, Ferraro PM, Unwin R. Inflammation and kidney stones: cause and effect? Curr Opin Urol. 2023;33:129–35. 10.1097/mou.0000000000001066. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

