Epigenetic regulation of cancer stemness
- PMID: 40744921
- PMCID: PMC12314033
- DOI: 10.1038/s41392-025-02340-6
Epigenetic regulation of cancer stemness
Abstract
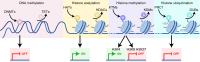
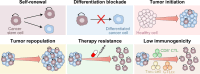
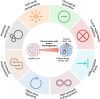
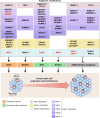
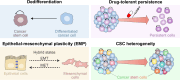
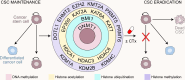
Gene expression is finely controlled by the abundance and activation status of transcription factors and their regulators, as well as by a number of reversible modifications of DNA and histones that are commonly referred to as epigenetic marks. Such alterations (i.e., methylation, acetylation, and ubiquitination) are catalyzed by an array of dedicated enzymes with antagonistic activity, including methyltransferases and demethylases, acetyltransferases and deacetylases, as well as ubiquitin ligases and deubiquitinating enzymes. The epigenetic control of transcription is critical not only for embryonic and postembryonic development but also for the preservation of homeostasis in all adult tissues. In line with this notion, epigenetic defects have been associated with a variety of human disorders, including (but not limited to) congenital conditions as well as multiple hematological and solid tumors. Here, we provide an in-depth discussion of the impact of epigenetic alterations on cancer stemness, i.e., the ability of a small population of poorly differentiated malignant cells to (1) self-renew while generating a more differentiated progeny, and (2) exhibit superior tumor initiating/repopulating potential along with exceptional plasticity and improved resistance to environmental and therapy-elicited stress. Moreover, we critically evaluate the potential and limitations of targeting epigenetic modifiers as a means to eradicate cancer stem cells for therapeutic purposes.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: ME is/has been holding research contracts with Ferrer International and Incyte and receives personal fees from Quimatryx (outside the scope of this work). LG is/has been holding research contracts with Lytix Biopharma, Promontory, and Onxeo; has received consulting/advisory honoraria from Boehringer Ingelheim, AstraZeneca, OmniSEQ, Onxeo, The Longevity Labs, Inzen, Imvax, Sotio, Promontory, Noxopharm, EduCom, and the Luke Heller TECPR2 Foundation; and holds Promontory stock options. The other authors have no conflicts of interest to declare.
Figures
References
-
- Cramer, P. Organization and regulation of gene transcription. Nature573, 45–54 (2019). - PubMed
-
- Lambert, S. A. et al. The human transcription factors. Cell172, 650–665 (2018). - PubMed
-
- Allis, C. D. & Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet.17, 487–500 (2016). - PubMed
-
- Millán-Zambrano, G., Burton, A., Bannister, A. J. & Schneider, R. Histone post-translational modifications-cause and consequence of genome function. Nat. Rev. Genet.23, 563–580 (2022). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

