vPro-MS enables identification of human-pathogenic viruses from patient samples by untargeted proteomics
- PMID: 40744923
- PMCID: PMC12314097
- DOI: 10.1038/s41467-025-62469-4
vPro-MS enables identification of human-pathogenic viruses from patient samples by untargeted proteomics
Abstract
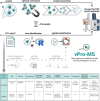
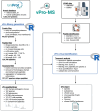
Viral infections are commonly diagnosed by the detection of viral genome fragments or proteins using targeted methods such as PCR and immunoassays. In contrast, metagenomics enables the untargeted identification of viral genomes, expanding its applicability across a broader spectrum. In this study, we introduce proteomics as a complementary approach for the untargeted identification of human-pathogenic viruses from patient samples. The viral proteomics workflow (vPro-MS) is based on an in-silico derived peptide library covering the human virome in UniProtKB (331 viruses, 20,386 genomes, 121,977 peptides). A scoring algorithm (vProID score) is developed to assess the confidence of virus identification from proteomics data ( https://github.com/RKI-ZBS/vPro-MS ). In combination with diaPASEF-based data acquisition, this workflow enables the analysis of up to 60 samples per day. The specificity is determined to be >99,9% in an analysis of 221 plasma, swab and cell culture samples covering 17 different viruses. The sensitivity of this approach for the detection of SARS-CoV-2 in nasopharyngeal swabs corresponds to a PCR cycle threshold of 27 with comparable quantitative accuracy to metagenomics. vPro-MS enables the integration of untargeted virus identification in large-scale proteomic studies of biofluids such as human plasma to detect previously undiscovered virus infections in patient specimens.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
The effect of sample site and collection procedure on identification of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Dec 16;12(12):CD014780. doi: 10.1002/14651858.CD014780. Cochrane Database Syst Rev. 2024. PMID: 39679851 Free PMC article.
-
Evaluation of the Seegene Allplex™ RV master assay for one-step simultaneous detection of eight respiratory viruses in nasopharyngeal specimens.J Virol Methods. 2025 Jan;331:115042. doi: 10.1016/j.jviromet.2024.115042. Epub 2024 Oct 9. J Virol Methods. 2025. PMID: 39384158
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Laboratory-based molecular test alternatives to RT-PCR for the diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Oct 14;10(10):CD015618. doi: 10.1002/14651858.CD015618. Cochrane Database Syst Rev. 2024. PMID: 39400904
-
Antibody tests for identification of current and past infection with SARS-CoV-2.Cochrane Database Syst Rev. 2022 Nov 17;11(11):CD013652. doi: 10.1002/14651858.CD013652.pub2. Cochrane Database Syst Rev. 2022. PMID: 36394900 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

