A founder BRCA1 exonic duplication involving breakpoint in T2T reference genome-specific region results in constitutional fusion transcript
- PMID: 40744942
- PMCID: PMC12314049
- DOI: 10.1038/s41525-025-00517-0
A founder BRCA1 exonic duplication involving breakpoint in T2T reference genome-specific region results in constitutional fusion transcript
Erratum in
-
Author Correction: A founder BRCA1 exonic duplication involving breakpoint in T2T reference genome-specific region results in constitutional fusion transcript.NPJ Genom Med. 2025 Sep 2;10(1):62. doi: 10.1038/s41525-025-00522-3. NPJ Genom Med. 2025. PMID: 40897690 Free PMC article. No abstract available.
Abstract
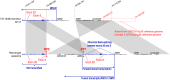
Pathogenicity assessment of genetic variants is the cornerstone of genetic counselling. Copy gains of exons are challenging, as pathogenicity depends on the localization of the additional exons. Eight patients form six families carried copy gains of BRCA1 exons 8-20. For appropriate characterization, long-read sequencing aligned on three distinct reference genome assemblies, optical genomic mapping, short-read and long-read RNA sequencing were performed. All patients shared the same pathogenic structural variant, involving a large segment located downstream in the genome. One breakpoint occurred in a region incorrectly annotated in GRCh37/hg19 and GRCh38/hg38. Alignment to the T2T-CHM13/hs1 assembly was therefore necessary for accurate characterization. This rearrangement caused various BRCA1 transcriptomic abnormalities: back-splicing, forward genomic strand transcription by insertion of an ectopic promoter, fusion transcripts with the "Next to BRCA1" gene 1 (NBR1). Our findings underscore the need to combine advanced technologies with the latest genome references to resolve complex rearrangements with significant medical implications.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Kuchenbaecker, K. B. et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA317, 2402–2416 (2017). - PubMed
-
- Brandt, T. et al. Adapting ACMG/AMP sequence variant classification guidelines for single-gene copy number variants. Genet. Med.22, 336–344 (2020). - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

