Stress granule clearance mediated by V-ATPase-interacting protein NCOA7 mitigates ovarian aging
- PMID: 40745099
- PMCID: PMC12350179
- DOI: 10.1038/s43587-025-00927-w
Stress granule clearance mediated by V-ATPase-interacting protein NCOA7 mitigates ovarian aging
Abstract
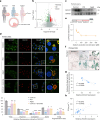
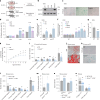
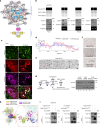
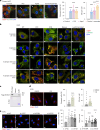
Reproductive longevity is essential for female fertility and healthy aging; however, the role of stress response, especially stress granule accumulation, in ovarian aging remains elusive and interventions are lacking. Here, we identified deleterious mutations and decreased expression of NCOA7, a stress-response protein related to granulosa cell senescence in women with physiological and pathological ovarian aging. NCOA7 deletion accelerates oxidative stress-related cellular senescence, ovarian aging and fecundity decline in mice. Mechanistically, NCOA7 partitions into the stress granule containing G3BP1-V-ATPase and facilitates autophagic degradation of stress granules to relieve stress. Boosting granulophagy with rapamycin or lipid nanoparticle-based mRNA delivery of NCOA7 accelerates stress granule clearance, alleviating cellular senescence in human granulosa cells and delaying ovarian aging in mice. This study depicts a mechanism for ovarian resilience to stress and provides potential targets for therapeutic strategies to alleviate ovarian aging.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





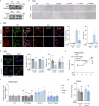
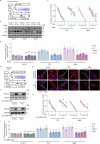
References
-
- Zhang, W., Qu, J., Liu, G. H. & Belmonte, J. C. I. The ageing epigenome and its rejuvenation. Nat. Rev. Mol. Cell Biol.21, 137–150 (2020). - PubMed
-
- Laisk, T. et al. Demographic and evolutionary trends in ovarian function and aging. Hum. Reprod. Update25, 34–50 (2019). - PubMed
-
- Jiao, X., Ke, H., Qin, Y. & Chen, Z. J. Molecular genetics of premature ovarian insufficiency. Trends Endocrinol. Metab29, 795–807 (2018). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

