Queuosine is incorporated into precursor tRNA before splicing
- PMID: 40745156
- PMCID: PMC12313893
- DOI: 10.1038/s41467-025-62220-z
Queuosine is incorporated into precursor tRNA before splicing
Abstract
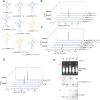
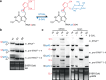
Each newly transcribed tRNA molecule must undergo processing and receive modifications to become functional. Queuosine (Q) is a tRNA modification present at position 34 of four tRNAs with "GUN" anticodons. Among these, the precursor of tRNATyr carries an intronic sequence within the anticodon loop that is removed by an essential non-canonical splicing event. The functional and temporal coupling between tRNA-splicing and Q-incorporation remains elusive. Here, we demonstrate in vitro and in vivo that intron-containing precursors of tRNATyr are modified with Q or with the Q-derivative galactosyl-queuosine (galQ) before being spliced. We show that this order of events is conserved in mouse, human, flies and worms. Using single particle cryo-EM, we confirm that pre-tRNATyr is a bona fide substrate of the QTRT1/2 complex, which catalyzes the incorporation of Q into the tRNA. Our results elucidate the hierarchical interplay that coordinates Q-incorporation and splicing in eukaryotic tRNAs, providing a relevant but unappreciated aspect of the cellular tRNA maturation process.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Ivanov, P. Ribosomes unexpectedly moonlight as activators of angiogenin enzyme. Nature630, 568–569 (2024). - PubMed
MeSH terms
Substances
Grants and funding
- 439669440 TRR319 RMaP TP A06/Deutsche Forschungsgemeinschaft (German Research Foundation)
- TU5371-2/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 442512666/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 439669440 TRR319 RMaP TP C03/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 101001394/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
LinkOut - more resources
Full Text Sources

