Integrated transcriptomic and functional modeling reveals AKT and mTOR synergy in colorectal cancer
- PMID: 40745175
- PMCID: PMC12313885
- DOI: 10.1038/s41598-025-08649-0
Integrated transcriptomic and functional modeling reveals AKT and mTOR synergy in colorectal cancer
Abstract
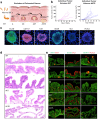
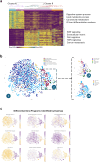
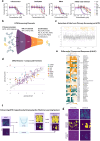
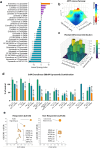
Colorectal cancer (CRC) treatment remains challenging due to genetic heterogeneity and resistance mechanisms. To address this, we developed a drug discovery pipeline using patient-derived primary CRC cultures with diverse genomic profiles. These cultures closely resemble certain molecular characteristics of primary and metastatic CRC, highlighting their promise as a translational platform for therapeutic evaluation. Importantly, our engineered model and patient-derived cells reflect the complexity and heterogeneity of primary tumors, not observed with standard immortalized cell lines, offering a more clinically relevant system, although further validation is needed. High-throughput screening (HTS) of 4255 compounds identified 33 with selective efficacy against CRC cells, sparing normal, healthy epithelial cells. Among the tested combinations, everolimus (mTOR inhibitor) and uprosertib (AKT inhibitor) demonstrated promising synergy at clinically relevant concentrations, with favorable therapeutic windows confirmed across tested patient-derived cultures. Notably, this synergy, revealed through advanced models, might have been overlooked in traditional immortalized cell lines, highlighting the translational advantage of patient-derived systems. Furthermore, the integration of machine learning into the HTS pipeline significantly improved scalability, cost-efficiency, and predictive accuracy. Our findings underscore the potential of patient-derived materials combined with machine learning-enhanced drug discovery to advance personalized therapies. Specifically, mTOR-AKT inhibition emerges as a promising strategy for CRC treatment, paving the way for more effective and targeted therapeutic approaches.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: MD, EZ, JS, MS, JS, OD, AK, IW, MC, BL, KS, JK, OB, ASW, KP, JD, KB, TR, AT, KB, AM are/were employees of Ryvu Therapeutics and some of the authors are shareholders in Ryvu Therapeutics. The other authors have no competing interests.
Figures


References
-
- Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer statistics. CA Cancer J. Clin/71(2021), 7–33. 10.3322/CAAC.21654 (2021). - PubMed
-
- Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.71, 209–249. 10.3322/CAAC.21660 (2021). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

