A framework for complex signal processing via synthetic biological operational amplifiers
- PMID: 40745425
- PMCID: PMC12314004
- DOI: 10.1038/s41467-025-62464-9
A framework for complex signal processing via synthetic biological operational amplifiers
Abstract
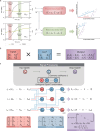
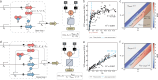
Engineering genetic circuits to process complex biological signals remains a significant challenge due to non-orthogonal signal responses that limit precise control. In this study, we introduce a framework that integrates orthogonal operational amplifiers (OAs) into standardized biological processes to enable efficient signal decomposition and amplification. By engineering σ/anti-σ pairs, varying ribosome binding site (RBS) strengths, and utilizing both open-loop and closed-loop configurations, we design scalable OAs that enhance the precision, adaptability, and signal-to-noise ratio of genetic circuits. Additionally, we present a prototype whole-cell biosensor capable of detecting transcriptional changes in response to growth conditions, enabling growth-state-responsive induction systems. These systems provide dynamic gene expression control without external inducers, offering significant advantages for metabolic engineering applications. We also apply our framework to mitigate crosstalk in multi-signal systems, ensuring independent control over each signal channel within complex biological networks. Our approach enhances synthetic biology systems by robust signal processing and precise dynamic regulation.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: Y.C., L.L., and W.C. are listed as inventors on a patent application (CN202510595826.5, PCT/CN2025/093793) related to the synthetic OA circuits described in this study. The remaining authors declare no competing interests.
Figures





References
-
- Wan, X. et al. Cascaded amplifying circuits enable ultrasensitive cellular sensors for toxic metals. Nat. Chem. Biol.15, 540–548 (2019). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

