Multiple analytical perspectives of mitochondrial genes in the context of preeclampsia: potential diagnostic markers
- PMID: 40746540
- PMCID: PMC12310717
- DOI: 10.3389/fimmu.2025.1595706
Multiple analytical perspectives of mitochondrial genes in the context of preeclampsia: potential diagnostic markers
Abstract
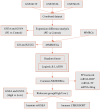
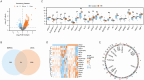
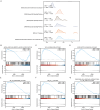
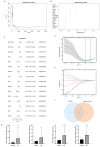
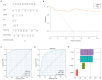
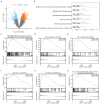
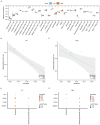
Preeclampsia(PE) is closely linked to adverse maternal and fetal outcomes. Given the pivotal roles of mitochondria in various human diseases and the limited research on their involvement in PE, this study identified biomarkers linked to mitochondrial metabolism in PE and their roles in its pathogenesis. Data from three datasets were integrated using the ComBat algorithm to mitigate batch effects. Differential expression analysis identified genes differentially expressed between PE cases and Control group. Cross-referencing these genes with mitochondrial energy metabolism-related genes (MMRGs) isolated mitochondrial energy metabolism-related differentially expressed genes (MMRDEGs). GO and KEGG analysis were performed to elucidate the functions of the MMRDEGs. A diagnostic model using Random Forest and logistic regression was validated by ROC curve analysis. mRNA expressions of OCRL, TPI1, GAPDH, and LDHA were quantified via qPCR. Immune characteristics were explored, and PPI, mRNA-miRNA, mRNA-TF and mRNA-RBP interaction networks were constructed. AlphaFold analyzed protein structures of OCRL, TPI1, GAPDH, and LDHA. A total of 1073 DEGs and 24 MMRDEGs were identified. OCRL, TPI1, GAPDH, and LDHA formed the diagnostic model, which were predominantly enriched in pyruvate metabolism, glycolysis, and ATP metabolism pathways. CIBERSORT highlighted immune cell composition variations between PE and Control groups. OCRL, TPI1, GAPDH, and LDHA exhibited increased mRNA expression levels in preeclamptic placentas. Therefore, MMRDEGs may play a critical role in the mechanism of oxidative stress and inflammatory response in PE by mediating metabolic regulation and immune modulation, potentially serving as diagnostic biomarkers associated with mitochondrial metabolism in preeclampsia.
Keywords: diagnostic model; immune cells infiltration; machine learning; mitochondria-related genes; preeclampsia.
Copyright © 2025 Li, Liu, Li, Zhao, Lv, Jiang and Zhao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

