Adjuvant combination and antigen multimerization shape neutralizing antibody and T cell responses to a SARS-CoV-2 RBD subunit vaccine
- PMID: 40746548
- PMCID: PMC12310705
- DOI: 10.3389/fimmu.2025.1610422
Adjuvant combination and antigen multimerization shape neutralizing antibody and T cell responses to a SARS-CoV-2 RBD subunit vaccine
Abstract
Introduction: The rapid development and deployment of multiple safe and effective COVID-19 vaccines were critical cornerstones of pandemic control. However, vaccine inequity and the emergence of new variants of concern (VOCs) highlighted major gaps in the global strategy to control SARS-CoV-2 infection. Despite the use of distinct platforms, most approved vaccines utilize the Spike protein as the main antigen due to its pivotal role in virus entry, mediated by the receptor binding domain (RBD). In this context, RBD stands out as a promising antigen for a subunit vaccine candidate, as it is the main target of neutralizing antibodies, has a well-established scalable production pipeline, and has proven safety. Approaches to enhance RBD immunogenicity encompass the addition of adjuvants and antigen multimerization.
Methods: In this study, we compared the immunogenic properties of the Wuhan RBD monomer and homodimer with an RBD heterotrimer formulation composed of the Delta, Beta and Gamma variants. We also screened different adjuvants to optimize both humoral and cellular immunity.
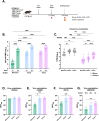
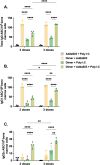
Results: Our results showed that immunization with the RBD dimer and trimer, in the presence of the adjuvant AddaS03, elicited a higher humoral response and a broader neutralization profile. Additionally, RBD-trimer immunization more efficiently inhibited viral replication in the lungs of mice challenged with the ancestral Wuhan strain compared to the monomer. We further optimized our vaccine formulation by combining the adjuvants AddaS03 and Poly I:C, which demonstrated a synergistic effect, integrating the potent humoral response induced by AddaS03 with the cellular Th1 skewing capacity of Poly I:C. The AddaS03+ Poly I:C mixture induced antibodies with higher affinity and an increased frequency of RBD-specific IgG2c-producing bone marrow plasma cells, highlighting the potential of this adjuvant combination to generate long-lived memory plasma cells. Additionally, we identified sequences within the RBD that induced specific IFNγ T cell responses. Peptide 12 (393-TNVYADSFVIRGDEVRQ-409) emerged as the immunodominant CD4 T cell epitope, whereas peptides 28 (505-YQPYRVVVLSFELLHAP-521) and 29 (512-VLSFELLHAPATVCGPK-528) successfully activated CD8 T cells.
Conclusions: These findings underscore that antigen multimerization and the strategic combination of adjuvants can significantly improve vaccine immunogenicity.
Keywords: RBD; SARS-CoV-2; adjuvant; recombinant protein; vaccine.
Copyright © 2025 Nunes, Silva, Apostolico, Daher, Marques, Yamamoto, Carvalho, de Castro-Amarante, Durigon, Wrenger, Janini, de Souza, Andreata-Santos, Maricato, Cunha-Neto, Kalil, Boscardin and Rosa.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

