Integrating single-cell regulatory atlas and multi-omics data for differential treatment response and multimodal predictive modeling in CDK 4/6 inhibitor-treated breast cancer
- PMID: 40746611
- PMCID: PMC12310732
- DOI: 10.3389/fonc.2025.1585574
Integrating single-cell regulatory atlas and multi-omics data for differential treatment response and multimodal predictive modeling in CDK 4/6 inhibitor-treated breast cancer
Abstract
Introduction: CDK4/6 inhibitors are cornerstone therapies for advanced HR+/HER2- breast cancer, yet treatment response heterogeneity remains a major clinical challenge. This study integrates single-cell regulatory landscapes with multi-omics data to decode resistance mechanisms and develop predictive biomarkers for CDK4/6 inhibitor response stratification.
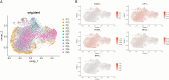
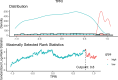
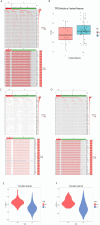
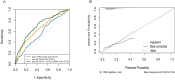
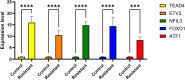
Methods: Single-cell RNA-seq data (GSE158724, n=14 samples) and bulk multi-omics profiles (TCGA-BRCA, n=1,059; GSE186901, n=90) were analyzed. Gene regulatory networks were reconstructed using SCENIC to identify resistance-specific regulons. The Tumor Prognostic Regulon Index (TPRI) was derived from five prognostic transcription factors and validated in independent cohorts. Experimental validation including qPCR of core TFs was performed in patient-derived samples. Multimodal predictive models integrating TPRI, differentially expressed genes, and miRNAs were developed using logistic regression, with performance assessed via ROC/AUC analysis.
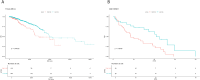
Results: We identified 86 resistance-associated regulons and established TPRI based on five prognostic TFs (ATF1, TEAD4, NFIL3, FOXO1, ETV3). TPRI significantly stratified patients into high/low-risk groups with differential overall survival and treatment response (Fisher's exact test P=0.0237). qPCR confirmed elevated expression of these TFs in resistant tumors (P<0.01). High-risk patients exhibited increased stemness indices (mRNAsi, P<2.2e-16) and mTOR pathway activation. The multimodal model (TPRI + top 30 DEGs + top 30 miRNAs) achieved superior prognostic accuracy (95%CI:0.6575-0.75).
Discussion: This study establishes TPRI as a novel biomarker for CDK4/6 inhibitor response prediction, validated through multi-omics integration and qPCR confirmation. The model provides actionable risk stratification, where high-risk patients may benefit from combinatorial mTOR-targeted therapies. Limitations include sample size constraints for methylation integration. Future studies should validate these findings in prospective clinical trials.
Keywords: CDK 4/6 inhibitors; TCGA; TPRI; breast cancer; prognostic model; single-cell sequencing; transcriptional regulation.
Copyright © 2025 Yan, Chen, Ran, Zhao, Huang and Yang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Cejalvo Andújar JM, Ayala de la Peña F, Margeli Vila M, Pascual J, Tolosa P, Pages C, et al. Optimizing therapeutic approaches for HR+/HER2- advanced breast cancer: clinical perspectives on biomarkers and treatment strategies post-CDK4/6 inhibitor progression. Cancer Drug Resist. (2025) 8:5. doi: 10.20517/cdr.2024.169, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

