Fungal Planet description sheets: 1781-1866
- PMID: 40746709
- PMCID: PMC12308287
- DOI: 10.3114/persoonia.2025.54.10
Fungal Planet description sheets: 1781-1866
Abstract
Novel species of fungi described in this study include those from various countries as follows: Argentina, Septoria reinamora on leaf spots of Mutisia spinosa. Australia, Cortinarius albofolliculus on mossy soil, Cortinarius descensoriformis among leaf litter, Cortinarius kaki among leaf litter, Cortinarius lissosporus in leaf litter, Cortinarius malogranatus in leaf litter, Cortinarius meletlac on soil in mixed forest, Cortinarius sebosioides in long decayed wood litter, Helicogermslita australiensis as an endophyte from healthy leaves of Archontophoenix cunninghamiana, Puccinia clemensiorum on culms of Eleocharis ochrostachys, Puccinia geethae on leaves of Cyperus brevifolius, Puccinia marjaniae on leaves of Nymphoides indica, Puccinia scleriae-rugosae on leaves of Scleria rugosa. Brazil, Dactylaria calliandrae on living leaf of Calliandra tweediei, Mucor cerradoensis from soil, Musicillium palmae on living leaves of unidentified palm species, Neodendryphiella agapanthi from stalks of Agapanthus praecox, Parafusicladium riodejaneiroanum on living leaves of native bamboo, Parapenidiella melastomatis on living leaves of unidentified Melastomataceae, Pararamichloridium ouropretoense on living leaves of unidentified Poaceae, Pentagonomyces endophyticus (incl. Pentagonomyces gen. nov.) as endophytic from roots of Musa acuminata, Polyschema endophytica from healthy roots of coffee plant, Purimyces endophyticus as root endophyte of Cattleya locatellii, Ramularia rhododendri on living leaves of Rhododendron sp., Staphylotrichum soli from soil, Trichoderma sexdentis from leaves inside a nest of the leaf-cutting ant Atta sexdens rubropilosa, Wiesneriomyces soli from soil. France, Cosmospora nemaniae on dead or effete stromata of Nemania cf. colliculosa, Inocybe alnobetulae in subalpine green alder stands, Stylonectria hygrophila on dead twigs of Betula pubescens. Germany, Coniochaeta corticalis from bark humus, Coniochaeta fermentaria from fermentation residues from biogas plants, Coniochaeta fibricola from softwood fibres, Coniochaeta weberae from bark humus, Inocybe canicularis on calcareous to more acidic soil with conifers. Iceland, Inocybe islandica associated with Dryas octopetala. India, Vishniacozyma indica on dead twigs. Iran, Botryotrichum lycii on rotten leaf of Lycium depressum. Italy, Cuphophyllus dolomiticus among Salix retusa, Salix reticulata and Dryas octopetala, Inocybe subentolomospora on moss with the presence of Alnus incana, Populus nigra and Salix spp. Malaysia, Catenulostroma pellitae on leaf spots of Eucalyptus pellita. Mexico, Colletotrichum mexicanus from fruit of Persea americana cv. Hass. New Caledonia (France), Cortinarius caeloculus, Cortinarius luteigemellus and Cortinarius perpensus on soil under Nothofagus aequilateralis. New Zealand, Cytospora braithwaitei on branch of Malus domestica. Pakistan, Callistosporium khalidii on humus soil, Entoloma lilacinum on litter in conifer forest, Laccaria decolorans on litter in broad-leaved subtropical forest. Poland, Pseudoneoconiothyrium modrzynanum from resin of Larix decidua ssp. polonica, Tuberculiforma enigmatica isolated from sooty mould community on Quercus robur leaves. Portugal, Clavulus hemisphaericus (incl. Clavulus gen. nov.) on mossy slopes and under Laurus leaf litter, Entoloma daegae on sandy, granitic soil, Hygrocybe aurantiocitrina under laurel forest, Hygrocybe sanguineolutea gregarious in laurel forest, Hygrocybe vulcanica on mossy areas of laurel forest areas, Pachyphlodes algarvensis on sandy soil under Cistus salvifolius, Quercus suber and Pinus pinea. South Africa, Amycosphaerella podalyriae on leaf of Podalyria calyptrata, Erythrobasidium eucalypti from the gut of Gonipterus sp., Letendraea goniomae on leaves of Gonioma kamassi, Pezicula brabeji and Sphaerulina brabeji on twigs of Brabejum stellatifolium, Stachybotrys conicosiae on dead flower head of Conicosia elongata, Talaromyces ignescens from soil. Spain, Cortinarius phaeobrunneus on soil under Quercus ilex and Q. faginea, Inocybe pini-halepensis among grass and fallen leaves of Pinus halepensis, Inocybe subporcorum in sandy soils under Quercus ilex subsp. ballota and Pinus pinaster, Mycena morenoi on dead leaves of Betula pubescens and Salix atrocinerea, Pachyphlodes iberica on clayey and loamy soil under Quercus ilex and Quercus rotundifolia, Ramariopsis coronata in laurel forest. Switzerland, Inocybe minata in a bog on very wet acidic soil with Salix spp. and Betula spp. Thailand, Hypocrella khonsanitii on scale insects (Coccidae), Petchiella hymenopterorum on hymenopteran pupae in the nest (Hymenoptera). Trinidad and Tobago, Neodevriesia maravalensis from office swab. Türkiye, Russula anatolica under Quercus vulcanica. UK, Paracylindrosporium dactylorhizae (incl. Paracylindrosporium gen. nov.) on leaf spots of Dactylorhiza sp., Niesslia hepworthiae and Niesslia libertiae on living leaves of Libertia grandiflora. Ukraine, Lichenohendersonia cetrariae on thallus of terricolous Cetraria aculeata. USA, Atromagnispora indianensis (incl. Atromagnispora gen. nov.) on submerged wood in a freshwater stream, Cytospora michiganensis from utility room (settle plate), Exophiala aeris from air (settle plate), Hongoboletus americanus from mixed pine-hardwood forest, Lorrainsmithia pennsylvanica from bedroom, air, Superstratomyces massachusettsanus from lyse buffer. Vietnam, Aspergillus halopiscium on dry marine anchovy Stolephorus commersonnii. Morphological and culture characteristics are supported by DNA barcodes. Citation: Crous PW, Catcheside DEA, Catcheside PS, Alfenas AC, Alfenas RF, Barreto RW, Lebel T, Balashov S, Broadbridge J, Jurjević Ž, De la Peña-Lastra S, Hoffmann R, Mateos A, Riebesehl J, Shivas RG, Soliz Santander FF, Tan YP, Altés A, Bandini D, Carriconde F, Cazabonne J, Czachura P, Gryta H, Eyssartier G, Larsson E, Pereira OL, Rigueiro-Rodríguez A, Wingfield MJ, Ahmad W, Bibi S, Denman S, Esteve-Raventós F, Hussain S, Illescas T, Luangsa-ard JJ, Möller L, Mombert A, Noisripoom W, Olariaga I, Pancorbo F, Paz A, Piątek M, Polman-Short C, Suárez E, Afshan NS, Ali H, Arzanlou M, Ayer F, Barratt J, Bellanger J-M, Bidaud A, Bishop-Hurley SL, Bohm M, Bose T, Campo E, Chau NB, Çolak ÖF, Cordeiro TRL, Cruz MO, Custódio FA, Couceiro A, Darmostuk V, Dearnaley JDW, de Azevedo Santiago ALCM, de Freitas LWS, de J Yáñez-Morales M, Domnauer C, Dentinger B, Dhileepan K, De Souza JT, Dovana F, Eberhardt U, Eisvand P, Erhard A, Fachada V, García-Martín A, Groenewald M, Hammerbacher A, Harms K, Haroon S, Haqnawaz M, Henriques S, Hernández AJ, Jacobus LM, Jaen-Contreras D, Jangsantear P, Kaygusuz O, Knoppersen R, Kumar TKA, Lynch MJ, Mahiques R, Maraia GL, Marbach PAS, Mehrabi-Koushki M, Miller PR, Mongkolsamrit S, Moreau P-A, Oberlies NH, Oliveira JA, Orlovich D, Pérez-Méndez AS, Pinto A, Raja HA, Ramírez GH, Raphael B, Rodrigues A, Rodrigues H, Ramos DO, Safi A, Sarwar S, Saar I, Sánchez RM, Santana JS, Scrace J, Sales LS, Silva LNP, Stryjak-Bogacka M, Tacconi A, Thanh VN, Thomas A, Thuy NT, Toome M, Valdez-Carrazco JM, van Vuuren NI, Vasey J, Vauras J, Vila-Viçosa C, Villarreal M, Visagie CM, Vizzini A, Whiteside EJ, Groenewald JZ. (2025). Fungal Planet description sheets: 1781-1866. Persoonia 54: 327-587. doi: 10.3114/persoonia.2025.54.10.
Keywords: ITS nrDNA barcodes; LSU; new taxa; systematics.
© 2025 Naturalis Biodiversity Center & Westerdijk Fungal Biodiversity Institute.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures





























































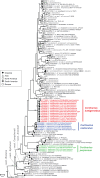

































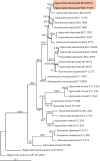

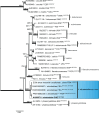







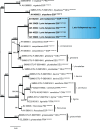








































References
-
- Anon (2016). Fact Sheet – Cyperus brevifolius. In: Weeds of Australia. https://keyserver.lucidcentral.org/weeds/data/media/Html/cyperus_brevifo... [accessed 3 Sep. 2024].
-
- Ariyawansa HA, Tanaka K, Thambugala KM, et al. (2014). A molecular phylogenetic reappraisal of the Didymosphaeriaceae (= Montagnulaceae). Fungal Diversity 68: 69–104. 10.1007/s13225-014-0305-6 - DOI
-
- Armada F, Bellanger J-M, Moreau P-A. (2024). Champignons de la zone alpine. Fédération Mycologique et Botanique Dauphiné-Savoie. Annemasse. France.
-
- Arnolds E. (1990). Hygrocybe (Fr.) Kumm., Flora Agaricina Neerlandica vol. 2, A.A. Balkema. Rotterdam.
Associated data
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
