Machine learning-enhanced discovery of tsRNA-mRNA regulatory networks: identifying novel diagnostic biomarkers and therapeutic targets in breast cancer
- PMID: 40746721
- PMCID: PMC12310640
- DOI: 10.3389/fphar.2025.1640192
Machine learning-enhanced discovery of tsRNA-mRNA regulatory networks: identifying novel diagnostic biomarkers and therapeutic targets in breast cancer
Abstract
Background: Transfer RNA-derived small RNAs (tsRNAs) represent an emerging class of regulatory molecules with potential as cancer biomarkers. However, their diagnostic utility and regulatory mechanisms in breast cancer remain poorly characterized. This study integrates machine learning algorithms with traditional molecular biology approaches to identify tsRNA-based diagnostic signatures and their downstream targets.
Methods: We analyzed miRNA-seq data from 103 matched tumor-normal pairs from TCGA-BRCA as the discovery cohort and GSE117452 as validation. tsRNA profiles were extracted using a custom bioinformatics pipeline. Random forest algorithm was employed to develop a diagnostic model. Correlation analysis and RNAhybrid were used to identify tsRNA-mRNA regulatory relationships. Comprehensive multi-omics analyses including survival, immune infiltration, drug sensitivity, and pathway enrichment were performed for identified targets. Functional validation was conducted in breast cancer cell lines.
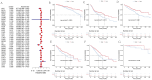
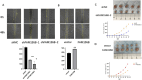
Results: We identified 297 differentially expressed tsRNAs and developed a four-tsRNA signature (tRF-21-FSXMSL73E, tRF-20-XSXMSL73, tRF-23-FSXMSL730H, tRF-23-YJE76INB0J) achieving AUC of 0.98 in discovery and 0.82 in validation cohorts. tRF-21-FSXMSL73E showed strong correlation with FAM155B expression. Pan-cancer analysis revealed FAM155B overexpression in multiple malignancies with prognostic significance. FAM155B correlated with immune infiltration, drug resistance, and activation of oncogenic pathways. Functional studies confirmed FAM155B promotes breast cancer proliferation and migration.
Conclusion: Our machine learning approach successfully identified a robust tsRNA diagnostic signature and uncovered the tsRNA-FAM155B regulatory axis as a novel therapeutic target. This integrated methodology provides a framework for accelerating biomarker discovery by combining computational prediction with traditional validation, advancing precision medicine in breast cancer.
Keywords: FAM155B; biomarker discovery; breast cancer; machine learning; tsRNA.
Copyright © 2025 Ma, Wang, Yuan, Wang, Li, Zhao and Zhao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures












Similar articles
-
A novel tsRNA, tRF-33-6978WPRLXN4V0O inhibits breast cancer development via regulating PTEN/AKT pathway in BHLHA15-mediated manner.J Mol Med (Berl). 2025 Sep;103(9):1135-1148. doi: 10.1007/s00109-025-02566-5. Epub 2025 Jul 28. J Mol Med (Berl). 2025. PMID: 40719768
-
Deciphering Shared Gene Signatures and Immune Infiltration Characteristics Between Gestational Diabetes Mellitus and Preeclampsia by Integrated Bioinformatics Analysis and Machine Learning.Reprod Sci. 2025 Jun;32(6):1886-1904. doi: 10.1007/s43032-025-01847-1. Epub 2025 May 15. Reprod Sci. 2025. PMID: 40374866
-
Plasma exosomal tsRNA may be used as a marker for differential diagnosis of benign and malignant pulmonary nodules.J Transl Med. 2025 Jul 1;23(1):712. doi: 10.1186/s12967-025-06737-3. J Transl Med. 2025. PMID: 40597381 Free PMC article.
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
-
Artificial intelligence for diagnosing exudative age-related macular degeneration.Cochrane Database Syst Rev. 2024 Oct 17;10(10):CD015522. doi: 10.1002/14651858.CD015522.pub2. Cochrane Database Syst Rev. 2024. PMID: 39417312
References
LinkOut - more resources
Full Text Sources

