Dose escalation study of the HLA-A2-WT1 CD3 bispecific antibody RO7283420 in relapsed/refractory acute myeloid leukemia
- PMID: 40746940
- PMCID: PMC12311517
- DOI: 10.1016/j.bneo.2025.100110
Dose escalation study of the HLA-A2-WT1 CD3 bispecific antibody RO7283420 in relapsed/refractory acute myeloid leukemia
Abstract
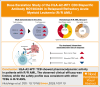
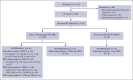
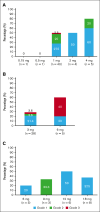
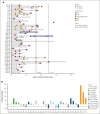
A novel T-cell bispecific antibody (TCB), RO7283420, engaging CD3 and the HLA-A2-Wilms tumor protein 1 complex, was evaluated in this phase 1 study to characterize safety and tolerability, determine the maximum tolerated dose (MTD), and recommend a phase 2 dose for patients with relapsed/refractory acute myeloid leukemia in 2 groups: hematologic (group I, n = 57) and molecular (group 2, n = 5) relapse. In group I, 51 received RO7283420 intravenously (IV) and 6 subcutaneously. The IV doses ranged from 0.15-4 mg (flat; n = 13), 3-18 mg (step-up; n = 34) every 3 weeks, or 9 mg weekly (step-up; n = 4). The MTD was 1/3/12 mg every 3 weeks. The most frequent adverse event in the overall population was cytokine release syndrome (61.3%) with grade ≥3 recorded in 9.7% of patients. Twelve dose-limiting toxicities were reported in 11 patients and 12 (19.4%) grade 5 adverse events, including 1 hemophagocytic lymphohistiocytosis case related to RO7283420. Among the 42 efficacy-evaluable IV patients in group I, 4.8% achieved complete remission (CR), and 2.4% achieved CR with incomplete hematologic recovery. RO7283420 induced pharmacodynamic changes in peripheral blood (PB) at doses ≥1 mg, including significant T-cell activation and expansion in the PB and bone marrow (BM). Significant associations were found between blast reduction and baseline immunophenotype, including lower regulatory T cells and higher non-exhausted CD8+ T cells in BM. Although dose escalation was discontinued because of limited efficacy and lack of an exposure-BM response relationship, the observed pharmacodynamics underscore the promising potential of this class of TCBs targeting intracellular antigens. This trial was registered at www.clinicaltrials.gov as #NCT04580121.
© 2025 American Society of Hematology. Published by Elsevier Inc. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: M.H. reports receiving research support for the institution from AbbVie, AstraZeneca, Bristol Myers Squibb (BMS)/Celgene, Genentech, Genmab, Incyte, Johnson & Johnson, Merck, Novartis, Roche, and Takeda; and receiving personal honoraria from AbbVie, AstraZeneca, Genmab, Johnson & Johnson, Merck, Roche, and Takeda. K.K. reports being an employee of and owning shares in F. Hoffmann-La Roche. P.M. reports receiving research support from AbbVie, Astellas, BMS, Daiichi Sankyo, Janssen, Novartis, Pfizer, and Teva; serving a consultancy role for Astellas, BMS, Daiichi Sankyo, and Glycomimetics; participating in speakers’ bureaus for AbbVie, Astellas, BMS, Daiichi Sankyo, Incyte, Janssen, Novartis, Pfizer, Sanofi, Servier, and Teva; and serving on advisory boards for AbbVie, Astellas, BMS, Daiichi Sankyo, Incyte, Janssen, Novartis, Pfizer, Sanofi, Servier, Syndax, and Teva. A.S. reports serving on advisory boards for Bayer, BMS, Eisai, Gilead, Merck Sharp & Dohme (MSD), Pfizer, and Servier; serving in a consultancy role for Incyte and Sanofi; and participating in speakers’ bureaus for AbbVie, Amgen, ArQule, AstraZeneca, Bayer, BeiGene, BMS/Celgene, Eisai, Gilead, Eli Lilly, MSD, Novartis, Pfizer, Roche, Sandoz, Servier, and Takeda. H.-A.H reports receiving a honorarium, travel support, and consultancy fees from Roche. S. Vives reports receiving travel support, accommodation, and expenses from AbbVie, Astellas, Jazz Pharmaceuticals, Pfizer, and Servier; receiving research funding from Astellas; and serving as a consultant or in an advisory role (without honoraria) for AbbVie, Astellas, Jazz Pharmaceuticals, Pfizer, and Servier. T.-Y.C. reports serving in a consultancy role for AbbVie, Amgen, BMS/Celgene, Janssen, and Sanofi. S. Garciaz reports serving in a consultancy or advisory role for AbbVie, Astellas, BMS/Celgene, Jazz Pharmaceuticals, and Servier; and receiving travel grants from Gilead. O.S.G. reports receiving honoraria from AbbVie, BMS, and Jazz Pharmaceuticals; serving in a consultancy role for AbbVie, BMS, and Astellas; serving on advisory boards for BMS and Novartis; and receiving travel support from Jazz Pharmaceuticals and Servier. S.-P.Y. received advisory board and/or lecture fees from AbbVie, Amgen, Astellas, Astex Pharmaceuticals, AstraZeneca, BeiGene, BMS, Janssen Pharmaceuticals, Novartis, Sanofi, and Takeda. K.Y. reports serving in a consultancy role for BMS/Celgene, F. Hoffmann-La Roche, GlaxoSmithKline (GSK), Jazz Pharmaceuticals, Novartis, Pfizer, Shattuck Labs, Taiho Oncology, and Takeda; receiving research funding from Astex Pharmaceuticals, F. Hoffmann-La Roche/Genentech, Forma Therapeutics, Geron Corporation, Gilead Sciences, Janssen, Jazz Pharmaceuticals, Novartis, and Treadwell Therapeutics; and receiving honoraria from AbbVie, Novartis, and Taiho. J.E. reports receiving research support from AbbVie, Jazz Pharmaceuticals, and Novartis; and providing scientific advice to AbbVie, Amgen, Astellas, BMS, Jazz Pharmaceuticals, Novartis, and Pfizer. A.B. reports receiving honoraria from AbbVie, Amgen, Astellas, Novartis, Pfizer, and Takeda; serving in an advisory role for Senti Bio and Shoreline; and participating in speakers’ bureaus for Amgen and BMS. S.F. reports serving in a consultancy role for Gilead; receiving research funding from Amgen; receiving honoraria from Amgen, BMS, Gilead, Novartis, Pfizer, and Servier; participating in speakers’ bureaus for Amgen, BMS, Gilead, Novartis, Pfizer, and Servier; and serving on the board of directors or advisory committees for Amgen, BMS, Gilead, Novartis, Pfizer, and Servier. A.C.B. reports being an employee of Roche Diagnostic GmbH and owning shares in F. Hoffmann-La Roche. J.A. reports being an employee of and owning shares in F. Hoffmann-La Roche and biotechnology funds, which may indirectly include pharmaceutical company shares. M. Sun, Y.M.M., G.S., H.Y., H.H., T.B., S.S., and S.N. report being employees of and owning shares in F. Hoffmann-La Roche. T.R. and S. Vauleon report being employees of F. Hoffmann-La Roche. C.K. reports being an employee of F. Hoffmann-La Roche at the time of the study and owning shares in F. Hoffmann-La Roche; and receiving patents/royalties from F. Hoffmann-La Roche. M.R. reports being an employee of F. Hoffmann-La Roche at the time of the study and owning shares in F. Hoffmann-La Roche. N.K. reports being an employee of and owning shares in F. Hoffmann-La Roche; and reports owning shares in Jazz Pharmaceuticals. M. Subklewe reports receiving research support from Amgen, BMS/Celgene, Gilead/Kite, Janssen, Miltenyi Biotec, Molecular Partners, Novartis, Roche, Seagen, Takeda; participating in speakers’ bureaus for AstraZeneca, BMS, Gilead/Kite, GSK, Janssen, Novartis, Pfizer, Roche, Springer Healthcare; serving in a consultancy role or on an advisory board for AbbVie, Amgen, Autolus, AvenCell, BMS, CanCell Therapeutics, Genmab US, Gilead, Ichnos Sciences, Incyte Biosciences, Interius BioTherapeutics, Janssen, Miltenyi Biomedicine, Molecular Partners, Nektar Therapeutics, Novartis, Orbital Therapeutics, Pfizer, Roche, Sanofi, Scare, Takeda; and receiving travel support from Gilead, Pfizer, and Roche. The remaining authors declare no competing financial interests.
Figures



References
-
- Döhner H, Wei AH, Appelbaum F, et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood. 2022;140(12):1345–1377. - PubMed
-
- National Cancer Institute Surveillance, Epidemiology, and End Results Program Cancer stat facts: leukemia - acute myeloid leukemia (AML) https://seer.cancer.gov/statfacts/html/amyl.html
-
- Percival M-E, Estey E. Emerging treatments in acute myeloid leukemia: current standards and unmet challenges. Clin Adv Hematol Oncol. 2017;15(8):632–642. - PubMed
Publication types
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials

