E3 ubiquitin ligase FBXW11-mediated downregulation of S100A11 promotes sensitivity to PARP inhibitor in ovarian cancer
- PMID: 40747341
- PMCID: PMC12311512
- DOI: 10.1016/j.jpha.2025.101246
E3 ubiquitin ligase FBXW11-mediated downregulation of S100A11 promotes sensitivity to PARP inhibitor in ovarian cancer
Abstract
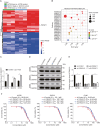
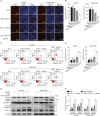
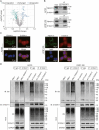
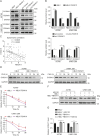
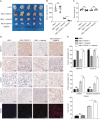
Resistance to poly adenosine diphosphate (ADP)-ribose polymerase inhibitor (PARPi) presents a considerable obstacle in the treatment of ovarian cancer. F-box and tryptophan-aspartic (WD) repeat domain containing 11 (FBXW11) modulates the ubiquitination of growth-and invasion-related factors in lung cancer, colorectal cancer, and osteosarcoma. The function of FBXW11 in PARPi therapy is still ambiguous. In this study, RNA sequencing (RNA-seq) showed that FBXW11 expression was raised in ovarian cancer cells that had been treated with PARPi. FBXW11 was abnormally expressed at low levels in high-grade serous ovarian cancer (HGSOC) tissues, and low levels of FBXW11 were associated with shorter overall survival (OS) and progression-free survival (PFS) in HGSOC patients. Overexpressing FBXW11 made ovarian cancer more sensitive to PARPi, while knocking down FBXW11 made it less sensitive. The four-dimensional (4D) label-free quantitative proteomic analysis revealed that FBXW11 targeted S100 calcium binding protein A11 (S100A11) and promoted its degradation through ubiquitination. The increased degradation of S100A11 led to less efficient DNA damage repair, which in turn contributed to increased PARPi-induced DNA damage. The role of FBXW11 in promoting PARPi sensitivity was also confirmed in xenograft mouse models. In summary, our study confirms that FBXW11 promotes the susceptibility of ovarian cancer cells to PARPi via affecting S100A11-mediated DNA damage repair.
Keywords: FBXW11; Ovarian cancer; PARPi resistance; S100A11.
© 2025 The Author(s).
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures



Similar articles
-
Poly(ADP-ribose) polymerase (PARP) inhibitors for the treatment of ovarian cancer.Cochrane Database Syst Rev. 2022 Feb 16;2(2):CD007929. doi: 10.1002/14651858.CD007929.pub4. Cochrane Database Syst Rev. 2022. PMID: 35170751 Free PMC article.
-
DNMT1 is required for efficient DSB repair and maintenance of replication fork stability, and its loss reverses resistance to PARP inhibitors in cancer cells.Oncogene. 2025 Jul;44(27):2283-2302. doi: 10.1038/s41388-025-03409-w. Epub 2025 Apr 15. Oncogene. 2025. PMID: 40234721
-
STING agonism overcomes STAT3-mediated immunosuppression and adaptive resistance to PARP inhibition in ovarian cancer.J Immunother Cancer. 2023 Jan;11(1):e005627. doi: 10.1136/jitc-2022-005627. J Immunother Cancer. 2023. PMID: 36609487 Free PMC article.
-
Controversies and clinical unknowns in the use of PARP inhibitors in ovarian cancer.Ther Adv Med Oncol. 2025 Jun 14;17:17588359251343973. doi: 10.1177/17588359251343973. eCollection 2025. Ther Adv Med Oncol. 2025. PMID: 40529205 Free PMC article. Review.
-
Combining EHMT and PARP Inhibition: A Strategy to Diminish Therapy-Resistant Ovarian Cancer Tumor Growth while Stimulating Immune Activation.Mol Cancer Ther. 2024 Sep 1;23(9):1332–1347. doi: 10.1158/1535-7163.MCT-23-0613. Epub 2024 May 8. Mol Cancer Ther. 2024. PMID: 38714351 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

