Screening and Verification COPD-OSA Overlap Syndrome Core Genes Using Bioinformatics
- PMID: 40747360
- PMCID: PMC12311238
- DOI: 10.2147/COPD.S528703
Screening and Verification COPD-OSA Overlap Syndrome Core Genes Using Bioinformatics
Abstract
Background: When obstructive sleep apnea (OSA) and chronic obstructive pulmonary disease (COPD) coexist in a patient, it is called overlap syndrome (OS). However, the molecular mechanisms underpinning OS are unclear. To address this, we explored potential OS mechanisms using bioinformatics.
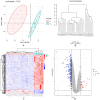
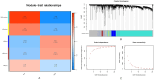
Methods: OSA and COPD gene expression datasets were obtained from the Gene Expression Omnibus (GEO) database. Differential expression and weighted gene co-expression network analyses (WGCNA) were performed to identify common differentially expressed genes (DEGs) in OSA and COPD, and perform functional enrichment analysis. DEGs were validated in an external COPD gene expression dataset using receiver operating characteristic (ROC) curves and box plots. Positive results were initially identified as core genes, and were then validated by analyzing core genes in healthy controls, patients with OSA alone and patients with OS using RT-qPCR.
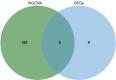
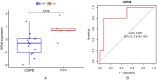
Results: Through differential expression gene analysis, 9 common DEGs for OSA and COPD were identified. Through WGCNA analysis, 128 common key module genes for OSA and COPD were identified. By taking the intersection of the identified 9 DEGs and the 128 common key module genes from WGCNA, 5 key genes were determined. Preliminary validation in the external gene expression dataset for COPD revealed that GRM8 was a potential hub gene for OS. Compared with the control group, the expression of GRM8 was significantly downregulated in the COPD group (P = 0.019). The diagnostic value was evaluated using the ROC curve, and the results showed that the AUC was 0.857 (95% CI: 0.614-1.000). Finally, RT-qPCR confirmed that the expression levels of GRM8 in OSA and OS were significantly lower than those in the healthy control group (P < 0.05), and it was a hub gene significantly associated with OS.
Conclusion: Our research identified hub gene that may provide new directions for further mechanistic research on OS.
Keywords: GRM8; bioinformatics; chronic obstructive pulmonary disease; glutamate metabotropic receptor 8; obstructive sleep apnea; overlap syndrome.
© 2025 Qiang et al.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






References
-
- Global Initiative for Chronic Obstructive Lung Disease (GOLD). Global Strategy for Prevention, Diagnosis and Management of COPD: 2025 Report. Bethesda: GOLD; 2025.
-
- Shawon MS, Perret JL, Senaratna CV, Lodge C, Hamilton GS, Dharmage SC. Current evidence on prevalence and clinical outcomes of co-morbid obstructive sleep apnea and chronic obstructive pulmonary disease: a systematic review. Sleep Med Rev. 2017;32:58–68. doi: 10.1016/j.smrv.2016.02.007 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

