Artifacts in spatial transcriptomics data: their detection, importance, prevalence, and prevention
- PMID: 40748322
- PMCID: PMC12315538
- DOI: 10.1093/bib/bbaf306
Artifacts in spatial transcriptomics data: their detection, importance, prevalence, and prevention
Abstract
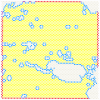
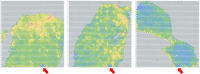
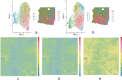
Data artifacts may induce errors in findings from any spatial transcriptomics platform. To provide protection from these errors, we have developed Border, Location, and edge Artifact DEtection (BLADE). BLADE is a novel collection of automated cross-platform statistical methods for detecting and removing three types of artifacts: (i) border effects, where total gene reads is modified at the border of the capture area; (ii) tissue edge effects, where total gene reads is modified at the edge of the tissue; (iii) location batch malfunctions, where there is a zone in the same location on all slides in a batch with substantially decreased sequencing depth. These artifacts are not mutually exclusive. BLADE has been applied to both Visium and CosMx data, and was used to evaluate our library of 37 10x Visium samples of liver and adipose tissue from humans and mice. Artifacts were found to be both common and impactful in those samples, indicating that artifact detection methods are critical for spatial transcriptomics quality control. Our BLADE software is publicly available.
Keywords: Visium; cellular senescence; modeling; quality control; spatial transcriptomics.
© The Author(s) 2025. Published by Oxford University Press.
Figures

References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

