SuperEdgeGO: Edge-supervised graph representation learning for enhanced protein function prediction
- PMID: 40749031
- PMCID: PMC12327639
- DOI: 10.1371/journal.pcbi.1013343
SuperEdgeGO: Edge-supervised graph representation learning for enhanced protein function prediction
Abstract
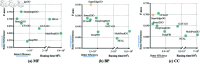
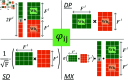
Understanding the functions of proteins is of great importance for deciphering the mechanisms of life activities. To date, there have been over 200 million known proteins, but only 0.2% of them have well-annotated functional terms. By measuring the contacts among residues, proteins can be described as graphs so that the graph leaning approaches can be applied to learn protein representations. However, existing graph-based methods put efforts in enriching the residue node information and did not fully exploit the edge information, which leads to suboptimal representations considering the strong association of residue contacts to protein structures and to the functions. In this article, we propose SuperEdgeGO, which introduces the supervision of edges in protein graphs to learn a better graph representation for protein function prediction. Different from common graph convolution methods that uses edge information in a plain or unsupervised way, we introduce a supervised attention to encode the residue contacts explicitly into the protein representation. Comprehensive experiments demonstrate that SuperEdgeGO achieves state-of-the-art performance on all three categories of protein functions. Additional ablation analysis further proves the effectiveness of the devised edge supervision strategy. The implementation of edge supervision in SuperEdgeGO resulted in enhanced graph representations for protein function prediction, as demonstrated by its superior performance across all the evaluated categories. This superior performance was confirmed through ablation analysis, which validated the effectiveness of the edge supervision strategy. This strategy has a broad application prospect in the study of protein function and related fields.
Copyright: © 2025 Zhang et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

