Neuronal Reg3β/macrophage TNF-α-mediated positive feedback signaling contributes to pain chronicity in a rat model of CRPS-I
- PMID: 40749060
- PMCID: PMC12315989
- DOI: 10.1126/sciadv.adu4270
Neuronal Reg3β/macrophage TNF-α-mediated positive feedback signaling contributes to pain chronicity in a rat model of CRPS-I
Abstract
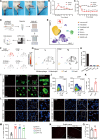
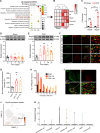
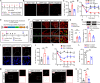
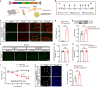
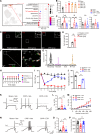
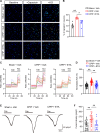
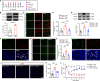
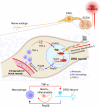
Complex regional pain syndrome type I (CRPS-I) develops after an initial injury. It causes prolonged pain that persists beyond the usual expected time for tissue healing. Mechanisms underlying pain chronicity of CRPS-I remain unknown. Here, we identified the presence of long-lasting infiltration of macrophages in local dorsal root ganglia (DRG) of a rat model of CRPS-I. We demonstrate that regenerating islet-derived 3β (Reg3β) is specifically produced by DRG neurons upon model establishment and functions as an important signaling molecule to drive proinflammatory macrophage infiltration in local DRG. Infiltrated macrophages produce TNF-α, which causes hyperexcitability of nociceptive DRG neurons and reciprocally promotes Reg3β overexpression and secretion from DRG neurons to recruit more macrophages. Our work reveals a positive feedback signaling conveyed by neuronal Reg3β/macrophage TNF-α that contributes to neuroinflammation in DRG, resulting in persistent pain in a rat model of CRPS-I. This finding provides insights into the neuroimmune interaction in local DRG that contributes to pain chronicity of CRPS-I.
Figures


References
-
- Ott S., Maihöfner C., Signs and symptoms in 1,043 patients with complex regional pain syndrome. J. Pain 19, 599–611 (2018). - PubMed
-
- Fassio A., Mantovani A., Gatti D., Rossini M., Viapiana O., Gavioli I., Benini C., Adami G., Pharmacological treatment in adult patients with CRPS-I: A systematic review and meta-analysis of randomized controlled trials. Rheumatology 61, 3534–3546 (2022). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

