Machine learning combining external validation to explore the immunopathogenesis of diabetic foot ulcer and predict therapeutic drugs
- PMID: 40749079
- PMCID: PMC12316216
- DOI: 10.1371/journal.pone.0328906
Machine learning combining external validation to explore the immunopathogenesis of diabetic foot ulcer and predict therapeutic drugs
Abstract
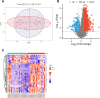
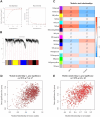
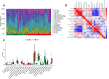
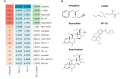
Diabetic foot ulcer (DFU) is a severe complication of diabetes, often leading to amputation due to poor wound healing and infection. The immune-related pathogenesis of DFU remains unclear, and therapeutic drugs are limited. This study aimed to explore the immune mechanisms of DFU and identify potential therapeutic drugs using machine learning and single-cell approaches. Through differential expression analysis of Gene Expression Omnibus (GEO) datasets, we identified 287 differentially expressed genes (DEGs), which were significantly enriched in IL-17 signaling and neutrophil chemotaxis pathways. Weighted gene co-expression network analysis (WGCNA) further pinpointed disease-associated modules containing 1,693 regulatory genes. Machine learning algorithms prioritized seven core genes-CCL20, CXCL13, FGFR2, FGFR3, PI3, PLA2G2A, and S100A8-with validation in an external dataset GSE147890 and single-cell sequencing revealing their predominant expression in neutrophils and keratinocytes. Immune infiltration analysis demonstrated significant dysregulation in DFU patients, characterized by elevated proportions of memory B cells, M0 macrophages, activated mast cells, and neutrophils. Potential therapeutic compounds were identified using the Connectivity Map database and tested through molecular docking and dynamics simulations. The study pinpointed selegiline, L-BSO, flunisolide, PP-30, and fluocinolone as promising therapeutic agents, offering new insights into the pathogenesis of diabetic foot ulcers (DFU) and potential therapeutic strategies.
Copyright: © 2025 Lu et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors declare no competing interests.
Figures







References
-
- Margolis DJ, Malay DS, Hoffstad OJ, Leonard CE, MaCurdy T, De Nava KL, et al. Incidence of diabetic foot ulcer and lower extremity amputation among medicare beneficiaries, 2006 to 2008: Data points# 2. Data points. 2011;2:4. - PubMed
-
- Forde H, Wrigley S, O’Murchadha LT, Cusack L, Casserly S, Moneley D. Five-year outcomes of patients attending a diabetic foot clinic in a tertiary referral centre. Irish Journal of Medical Science (1971-). 2020;189:511–5. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

