Structural convergence and water-mediated substrate mimicry enable broad neuraminidase inhibition by human antibodies
- PMID: 40750617
- PMCID: PMC12316982
- DOI: 10.1038/s41467-025-62339-z
Structural convergence and water-mediated substrate mimicry enable broad neuraminidase inhibition by human antibodies
Abstract
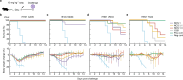
Influenza has been responsible for multiple global pandemics and seasonal epidemics and claimed millions of lives. The imminent threat of a panzootic outbreak of avian influenza H5N1 virus underscores the urgent need for pandemic preparedness and effective countermeasures, including monoclonal antibodies (mAbs). Here, we characterize human mAbs that target the highly conserved catalytic site of viral neuraminidase (NA), termed NCS mAbs, and the molecular basis of their broad specificity. Cross-reactive NA-specific B cells were isolated by using stabilized NA probes of non-circulating subtypes. We found that NCS mAbs recognized multiple NAs of influenza A as well as influenza B NAs and conferred prophylactic protections in mice against H1N1, H5N1, and influenza B viruses. Cryo-electron microscopy structures of two NCS mAbs revealed that they rely on structural mimicry of sialic acid, the substrate of NA, by coordinating not only amino acid side chains but also water molecules, enabling inhibition of NA activity across multiple influenza A and B viruses, including avian influenza clade 2.3.4.4b H5N1 viruses. Our results provide a molecular basis for the broad reactivity and inhibitory activity of NCS mAbs targeting the catalytic site of NA through substrate mimicry.
© 2025. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests: J.L., D.E., B.S.G., N.P.K., and M.K. are named inventors of a patent application describing engineered influenza neuraminidase antigens under publication number WO/2021/178621 filed by the University of Washington and the Department of Health and Human Services, USA. The remaining authors declare no competing interests.
Figures




Update of
-
Structural Convergence and Water-Mediated Substrate Mimicry Enable Broad Neuraminidase Inhibition by Human Antibodies.bioRxiv [Preprint]. 2024 Dec 3:2024.11.27.625426. doi: 10.1101/2024.11.27.625426. bioRxiv. 2024. Update in: Nat Commun. 2025 Aug 1;16(1):7068. doi: 10.1038/s41467-025-62339-z. PMID: 39677750 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- GNT1173433/Department of Health | National Health and Medical Research Council (NHMRC)
- ZIAAI005003/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R01 GM129325/GM/NIGMS NIH HHS/United States
- ZIA AI005003/ImNIH/Intramural NIH HHS/United States
- GNT2009711/Department of Health | National Health and Medical Research Council (NHMRC)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

