Synergistic H&E and IHC image analysis by AI predicts cancer biomarkers and survival outcomes in colorectal and breast cancer
- PMID: 40750675
- PMCID: PMC12317095
- DOI: 10.1038/s43856-025-01045-9
Synergistic H&E and IHC image analysis by AI predicts cancer biomarkers and survival outcomes in colorectal and breast cancer
Erratum in
-
Author Correction: Synergistic H&E and IHC image analysis by AI predicts cancer biomarkers and survival outcomes in colorectal and breast cancer.Commun Med (Lond). 2025 Oct 24;5(1):435. doi: 10.1038/s43856-025-01097-x. Commun Med (Lond). 2025. PMID: 41136588 Free PMC article. No abstract available.
Abstract
Background: Recent advancements in immunotherapy, particularly pembrolizumab, have shown promising results in treating metastatic colorectal cancer (CRC) and triple-negative breast cancer (TNBC). Accurate detection of predictive biomarkers, such as microsatellite instability (MSI)/mismatch repair deficiency (MMRd) and programmed death-ligand 1 (PD-L1), is key to efficacy of these treatments. Traditional methods like immunohistochemistry (IHC) and next-generation sequencing are effective but are labor intensive and require subjective interpretation.
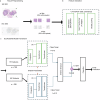
Methods: We developed a dual-modality transformer-based model for predicting MSI/MMRd and PD-L1 status using hematoxylin & eosin and IHC stained whole slide images. We evaluated the model using area under the receiver operating curve (AUROC). Time-on-treatment (TOT) and overall survival (OS) were derived from insurance claims and analyzed by Kaplan-Meier method. Hazard ratios (HR) were determined using the Cox proportional hazard model.
Results: Our AI framework achieves clinical-grade performance, with AUROC exceeding 0.97 for MSI/MMRd prediction in CRC and 0.96 for PD-L1 prediction in breast cancer. Patients with biomarker-positive model predictions demonstrated prolonged TOT and OS when treated with pembrolizumab. For breast cancer patients, the model's predictions were superior to PD-L1 IHC in stratifying patients with improved outcomes on pembrolizumab, suggesting a reevaluation of existing PD-L1 status thresholds.
Conclusions: This study promotes the integration of advanced AI tools in clinical pathology, aiming to enhance the precision and efficiency of cancer biomarker evaluation and offering a customizable framework for varied clinical scenarios. Our model enhances predictive accuracy, integrating features from both staining methods, and exhibits superior prognostic precision compared to current biomarker assessments.
Plain language summary
Current methods to identify cancer patients who will respond to specific immunotherapy treatments are labor intensive and require interpretation of markers in cancer tissue by clinicians. We developed a computer model that analyzes tumor tissue images to predict the status of key biomarkers that are used to select patients with colorectal cancer and triple-negative breast cancer for pembrolizumab treatment. We show that our model predicts statuses with high accuracy and identifies patients with improved outcomes on pembrolizumab. Clinical adoption of this tool could improve the precision and efficiency of cancer patient evaluation and aid clinical decision making.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: Y.C., N.L., M.C., E.A., M.R., M.A.R., J.X., A.H., L.D., J.R.R., H.G., M.O., D.S., and G.W.S. have a financial relationship as employees of Caris Life Sciences. The authors declare that they have no other known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





References
-
- Andre, T. et al. Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N. Engl. J. Med.383, 2207–2218 (2020). - PubMed
-
- Cortes, J. et al. Pembrolizumab plus chemotherapy in advanced triple-negative breast cancer. N. Engl. J. Med.387, 217–226 (2022). - PubMed
-
- Bartley, A. N. et al. Mismatch repair and microsatellite instability testing for immune checkpoint inhibitor therapy: guideline from the College of American Pathologists in collaboration with the Association for Molecular Pathology and Fight Colorectal Cancer. Arch. Pathol. Lab. Med.146, 1194–1210 (2022). - PubMed
-
- Sholl, L. M. et al. Programmed death ligand-1 and tumor mutation burden testing of patients with lung cancer for selection of immune checkpoint inhibitor therapies: guideline from the College of American Pathologists, Association for Molecular Pathology, International Association for the Study of Lung Cancer, Pulmonary Pathology Society, and LUNGevity Foundation. Arch. Pathol. Lab. Med. 10.5858/arpa.2023-0536-CP (2024). - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

